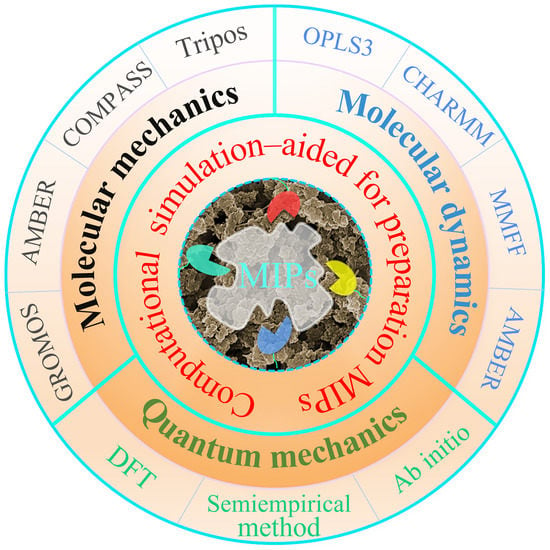
A Review on Molecularly Imprinted Polymers Preparation by Computational Simulation-Aided Methods
Abstract
:1. Introduction
2. Theoretical Methods of Computational Simulation for MIPs
2.1. MM Method
2.2. MD Method
2.3. QM Method
3. Computational Simulation and Design of New MIPs
4. Computational Simulation and MIP Identification Mechanism
5. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| MIP | Molecularly imprinted polymers |
| MM | Molecular mechanics |
| MD | Molecular dynamics |
| QM | Quantum mechanics |
| DFT | Density functional theory |
| OPLS | Optimized potentials for liquid simulations |
| CHARMM | Chemistry at Harvard macromolecular mechanics |
| MMFF | Merck molecular force field |
| Tripos | Tripos force field |
| COMPASS | Computer plan appraisal system for ship |
| GROMOS | Gromos force field |
| Ab initio | Latin term meaning “from the beginning” |
| B3LYP | Becke, three-parameter |
| GAFF | Generation amber force field |
| SDCI | Single and double excitation configuration interaction |
| CISD(T) | Configuration interactions with single and double substitutions |
| MCSCF | Multiconfiguration self-consistent field |
| AM1 | AM1 semiempirical method |
| PM3 | Semiempirical PM3 method |
| EHMO | Extended huckel molecular orbital theory |
| CNDO | Complete neglect of differential overlap method |
| NDDO | Neglect of diatomic differential overlap method |
References
- Pauling, L. A Theory of the structure and process of formation of antibodies. J. Am. Chem. Soc. 1940, 62, 2643–2657. [Google Scholar] [CrossRef]
- Wulff, G.; Sarhan, A.; Zabrocki, K. Enzyme-analogue built polymers and their use for the resolution of racemates. Tetrahedron Lett. 1973, 14, 4329–4332. [Google Scholar] [CrossRef]
- Kubo, T.; Nomachi, M.; Nemoto, K.; Sano, T.; Hosoya, K.; Tanaka, N.; Kaya, K. Chromatographic separation for domoic acid using a fragment imprinted polymer. Anal. Chim. Acta 2006, 577, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Sarafraz-Yazdi, A.; Razavi, N. Application of molecularly-imprinted polymers in solid-phase microextraction techniques. TrAC Trends Anal. Chem. 2015, 73, 81–90. [Google Scholar] [CrossRef]
- Song, Y.P.; Zhang, L.; Wang, G.N.; Liu, J.X.; Liu, J.; Wang, J.P. Dual-dummy-template molecularly imprinted polymer combining ultra performance liquid chromatography for determination of fluoroquinolones and sulfonamides in pork and chicken muscle. Food Control 2017, 82, 233–242. [Google Scholar] [CrossRef]
- Lian, Z.; Li, H.-B.; Wang, J. Experimental and computational studies on molecularly imprinted solid-phase extraction for gonyautoxins 2,3 from dinoflagellate Alexandrium minutum. Anal. Bioanal. Chem. 2016, 408, 5527–5535. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, O.S.; Bedwell, T.S.; Esen, C.; Garcia-Cruz, A.; Piletsky, S.A. Molecularly imprinted polymers in electrochemical and optical sensors. Trends Biotechnol. 2019, 37, 294–309. [Google Scholar] [CrossRef]
- Su, C.; Li, Z.; Zhang, D.; Wang, Z.; Zhou, X.; Liao, L.; Xiao, X. A highly sensitive sensor based on a computer-designed magnetic molecularly imprinted membrane for the determination of acetaminophen. Biosens. Bioelectron. 2020, 148, 111819. [Google Scholar] [CrossRef]
- Goud, K.Y.; Reddy, K.K.; Gobi, K.V. Development of highly selective electrochemical impedance sensor for detection of sub-micromolar concentrations of 5-Chloro-2,4-dinitrotoluene. J. Chem. Sci. 2016, 128, 763–770. [Google Scholar] [CrossRef]
- Gu, X.; Huang, J.; Zhang, L.; Zhang, Y.; Wang, C.-Z.; Sun, C.; Yao, D.; Li, F.; Chen, L.; Yuan, C.-S. Efficient discovery and capture of new neuronal nitric oxide synthase-postsynaptic density protein-95 uncouplers from herbal medicines using magnetic molecularly imprinted polymers as artificial antibodies. J. Sep. Sci. 2017, 40, 3522–3534. [Google Scholar] [CrossRef]
- Dong, Y.; Li, W.; Gu, Z.; Xing, R.; Ma, Y.; Zhang, Q.; Liu, Z. Inhibition of HER2-positive breast cancer growth by blocking the HER2 signaling pathway with HER2-glycan-imprinted nanoparticles. Angew. Chem. Int. Ed. 2019, 58, 10621–10625. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, Y.; Liu, Z.; Du, F.; Qin, G.; Li, G.; Hu, X.; Xu, Z.; Cai, Z. Supramolecularly imprinted polymeric solid phase microextraction coatings for synergetic recognition nitrophenols and bisphenol A. J. Hazard. Mater. 2019, 368, 358–364. [Google Scholar] [CrossRef]
- Li, S.; Yin, G.; Wu, X.; Liu, C.; Luo, J. Supramolecular imprinted sensor for carbofuran detection based on a functionalized multiwalled carbon nanotube-supported Pd-Ir composite and methylene blue as catalyst. Electrochim. Acta 2016, 188, 294–300. [Google Scholar] [CrossRef]
- Wang, S.; She, Y.; Hong, S.; Du, X.; Yan, M.; Wang, Y.; Qi, Y.; Wang, M.; Jiang, W.; Wang, J. Dual-template imprinted polymers for class-selective solid-phase extraction of seventeen triazine herbicides and metabolites in agro-products. J. Hazard. Mater. 2019, 367, 686–693. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, Y.; Liu, Z.; Hill, J.P.; Alowasheeir, A.; Xu, Z.; Xu, X.; Yamauchi, Y. Ultra-durable, multi-template molecularly imprinted polymers for ultrasensitive monitoring and multicomponent quantification of trace sulfa antibiotics. J. Mater. Chem. B 2021, 9, 3192–3199. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Hao, Y.; Zhao, S.; Zhang, L.; Cui, X.; Liu, D.; Tang, Y.; Zheng, Y. Novel magnetic multi-template molecularly imprinted polymers for specific separation and determination of three endocrine disrupting compounds simultaneously in environmental water samples. RSC Adv. 2014, 4, 56798–56808. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, L.; Dang, Y.; Chen, Z.; Zhang, R.; Li, Y.; Ye, B. A robust electrochemical sensing of molecularly imprinted polymer prepared by using bifunctional monomer and its application in detection of cypermethrin. Biosens. Bioelectron. 2019, 127, 207–214. [Google Scholar] [CrossRef]
- Marća, M.; Wieczorek, P.P. The preparation and evaluation of core-shell magnetic dummy-template molecularly imprinted polymers for preliminary recognition of the low-mass polybrominated diphenyl ethers from aqueous solutions. Sci. Total. Environ. 2020, 724, 138151. [Google Scholar] [CrossRef]
- Yuan, X.; Yuan, Y.; Gao, X.; Xiong, Z.; Zhao, L. Magnetic dummy-template molecularly imprinted polymers based on multi-walled carbon nanotubes for simultaneous selective extraction and analysis of phenoxy carboxylic acid herbicides in cereals. Food Chem. 2020, 333, 127540. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, H.-Y.; He, X.-W.; Li, W.-Y.; Zhang, Y.-K. Homochiral fluorescence responsive molecularly imprinted polymer: Highly chiral enantiomer resolution and quantitative detection of L-penicillamine. J. Hazard. Mater. 2021, 412, 125249. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, Y.; Liu, Z.; Zhao, X.; Wei, J.; Liu, H.; Si, X.; Xu, Z.; Cai, Z. Chiral molecularly imprinted polymeric stir bar sorptive extraction for naproxen enantiomer detection in PPCPs. J. Hazard. Mater. 2020, 392, 122251. [Google Scholar] [CrossRef]
- Lofgreen, J.E.; Ozin, G.A. Controlling morphology and porosity to improve performance of molecularly imprinted sol–gel silica. Chem. Soc. Rev. 2014, 43, 911–933. [Google Scholar] [CrossRef] [PubMed]
- Marć, M.; Kupka, T.; Wieczorek, P.P.; Namieśnik, J. Computational modeling of molecularly imprinted polymers as a green approach to the development of novel analytical sorbents. TrAC Trends Anal. Chem. 2018, 98, 64–78. [Google Scholar] [CrossRef]
- Paredes-Ramos, M.; Bates, F.; Rodríguez-González, I.; López-Vilariño, J.M. Computational approximations of molecularly imprinted polymers with sulphur based monomers for biological purposes. Mater. Today Commun. 2019, 20, 100526. [Google Scholar] [CrossRef]
- Cowen, T.; Karim, K.; Piletsky, S. Computational approaches in the design of synthetic receptors—A review. Anal. Chim. Acta 2016, 936, 62–74. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.S.; Pal, S.; Krupadam, R.J. Computational strategies for understanding the nature of interaction in dioxin imprinted nanoporous trappers. J. Mol. Recognit. 2015, 28, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Paredes-Ramos, M.; Sabín-López, A.; Peña-García, J.; Pérez-Sánchez, H.; López-Vilariño, J.M.; Sastre De Vicente, M.E. Computational aided acetaminophen—Phthalic acid molecularly imprinted polymer design for analytical determination of known and new developed recreational drugs. J. Mol. Graph. Model. 2020, 100, 107627. [Google Scholar] [CrossRef] [PubMed]
- Lai, W.; Zhang, K.; Shao, P.; Yang, L.; Ding, L.; Pavlostathis, S.G.; Shi, H.; Zou, L.; Liang, D.; Luo, X. Optimization of adsorption configuration by DFT calculation for design of adsorbent: A case study of palladium ion-imprinted polymers. J. Hazard. Mater. 2019, 379, 120791. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.V.; Dennison, S.R.; Archontis, G.; Reddy, S.M.; Hayes, J.M. Toward rational design of selective molecularly imprinted polymers (MIPs) for proteins: Computational and experimental studies of acrylamide based polymers for myoglobin. J. Phys. Chem. B 2019, 123, 5432–5443. [Google Scholar] [CrossRef]
- Xi, S.; Zhang, K.; Xiao, D.; He, H. Computational-aided design of magnetic ultra-thin dummy molecularly imprinted polymer for selective extraction and determination of morphine from urine by high-performance liquid chromatography. J. Chromatogr. A 2016, 1473, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Han, F.; Hu, Y.; Zheng, P.; Sheng, X.; Sun, H.; Song, W.; Lv, Y. Selective trace analysis of chloroacetamide herbicides in food samples using dummy molecularly imprinted solid phase extraction based on chemometrics and quantum chemistry. Anal. Chim. Acta 2012, 729, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Zhou, D.B.; Song, W.; Hu, Y.Y.; Lv, Y.N.; Ding, L.; Zheng, P.; Jia, X.Y.; Zhang, L.; Deng, X.J. Computational design and synthesis of molecular imprinted polymers for selective solid phase extraction of sulfonylurea herbicides. J. Chromatogr. A 2021, 1651, 462321. [Google Scholar] [CrossRef]
- Fizir, M.; Wei, L.; Muchuan, N.; Itatahine, A.; Mehdi, Y.A.; He, H.; Dramou, P. QbD approach by computer aided design and response surface methodology for molecularly imprinted polymer based on magnetic halloysite nanotubes for extraction of norfloxacin from real samples. Talanta 2018, 184, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Piletska, E.V.; Abd, B.H.; Krakowiak, A.S.; Parmar, A.; Pink, D.L.; Wall, K.S.; Wharton, L.; Moczko, E.; Whitcombe, M.J.; Karim, K.; et al. Magnetic high throughput screening system for the development of nano-sized molecularly imprinted polymers for controlled delivery of curcumin. Analyst 2015, 140, 3113–3120. [Google Scholar] [CrossRef] [PubMed]
- Bakas, I.; Ben Oujji, N.; Istamboulié, G.; Piletsky, S.; Piletska, E.; Ait-Addi, E.; Ait-Ichou, I.; Noguer, T.; Rouillon, R. Molecularly imprinted polymer cartridges coupled to high performance liquid chromatography (HPLC-UV) for simple and rapid analysis of fenthion in olive oil. Talanta 2014, 125, 313–318. [Google Scholar] [CrossRef]
- Karim, K.; Cowen, T.; Guerreiro, A.; Piletska, E.; Whitcombe, M. A protocol for the computational design of high affinity molecularly imprinted polymer synthetic receptors. Glob. J. Biotechnol. Biomater. Sci. 2017, 3, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Bakas, I.; Hayat, A.; Piletsky, S.; Piletska, E.; Chehimi, M.M.; Noguer, T.; Rouillon, R. Electrochemical impedimetric sensor based on molecularly imprinted polymers/sol–gel chemistry for methidathion organophosphorous insecticide recognition. Talanta 2014, 130, 294–298. [Google Scholar] [CrossRef]
- Abdin, M.J.; Altintas, Z.; Tothill, I.E. In silico designed nanoMIP based optical sensor for endotoxins monitoring. Biosens. Bioelectron. 2015, 67, 177–183. [Google Scholar] [CrossRef]
- Aftim, N.; Istamboulié, G.; Piletska, E.; Piletsky, S.; Calas-Blanchard, C.; Noguer, T. Biosensor-assisted selection of optimal parameters for designing molecularly imprinted polymers selective to phosmet insecticide. Talanta 2017, 174, 414–419. [Google Scholar] [CrossRef]
- Smolinska-Kempisty, K.; Ahmad, O.S.; Guerreiro, A.; Karim, K.; Piletska, E.; Piletsky, S. New potentiometric sensor based on molecularly imprinted nanoparticles for cocaine detection. Biosens. Bioelectron. 2017, 96, 49–54. [Google Scholar] [CrossRef]
- Esen, C.; Czulak, J.; Cowen, T.; Piletska, E.; Piletsky, S.A. Highly efficient abiotic assay formats for methyl parathion: Molecularly imprinted polymer nanoparticle assay as an alternative to enzyme-linked immunosorbent assay. Anal. Chem. 2019, 91, 958–964. [Google Scholar] [CrossRef] [PubMed]
- Sergeyeva, T.; Yarynka, D.; Piletska, E.; Lynnik, R.; Zaporozhets, O.; Brovko, O.; Piletsky, S.; El’Skaya, A. Fluorescent sensor systems based on nanostructured polymeric membranes for selective recognition of Aflatoxin B1. Talanta 2017, 175, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Qiu, C.; Xing, Y.; Yang, W.; Zhou, Z.; Wang, Y.; Liu, H.; Xu, W. Surface molecular imprinting on hybrid SiO 2 -coated CdTe nanocrystals for selective optosensing of bisphenol A and its optimal design. Appl. Surf. Sci. 2015, 345, 405–417. [Google Scholar] [CrossRef]
- He, Q.; Liang, J.-J.; Chen, L.-X.; Chen, S.-L.; Zheng, H.-L.; Liu, H.-X.; Zhang, H.-J. Removal of the environmental pollutant carbamazepine using molecular imprinted adsorbents: Molecular simulation, adsorption properties, and mechanisms. Water Res. 2020, 168, 115164. [Google Scholar] [CrossRef]
- Li, X.; Wan, J.; Wang, Y.; Ding, S.; Sun, J. Improvement of selective catalytic oxidation capacity of phthalates from surface molecular-imprinted catalysis materials: Design, mechanism, and application. Chem. Eng. J. 2021, 413, 127406. [Google Scholar] [CrossRef]
- Kong, Y.; Wang, N.; Ni, X.; Yu, Q.; Liu, H.; Huang, W.; Xu, W. Molecular dynamics simulations of molecularly imprinted polymer approaches to the preparation of selective materials to remove norfloxacin. J. Appl. Polym. Sci. 2016, 133. [Google Scholar] [CrossRef]
- Xu, W.; Wang, Y.; Huang, W.; Yu, L.; Yang, Y.; Liu, H.; Yang, W. Computer-aided design and synthesis of CdTe@SiO2 core-shell molecularly imprinted polymers as a fluorescent sensor for the selective determination of sulfamethoxazole in milk and lake water. J. Sep. Sci. 2017, 40, 1091–1098. [Google Scholar] [CrossRef]
- Silva, C.F.; Menezes, L.F.; Pereira, A.C.; Nascimento, C.S. Molecularly Imprinted Polymer (MIP) for thiamethoxam: A theoretical and experimental study. J. Mol. Struct. 2021, 1231, 129980. [Google Scholar] [CrossRef]
- Liu, R.; Li, X.; Li, Y.; Jin, P.; Qin, W.; Qi, J. Effective removal of rhodamine B from contaminated water using non-covalent imprinted microspheres designed by computational approach. Biosens. Bioelectron. 2009, 25, 629–634. [Google Scholar] [CrossRef]
- Yu, H.; Yao, R.; Shen, S. Development of a novel assay of molecularly imprinted membrane by design-based gaussian pattern for vancomycin determination. J. Pharm. Biomed. Anal. 2019, 175, 112789. [Google Scholar] [CrossRef] [PubMed]
- Prasad, B.B.; Kumar, A.; Singh, R. Molecularly imprinted polymer-based electrochemical sensor using functionalized fullerene as a nanomediator for ultratrace analysis of primaquine. Carbon 2016, 109, 196–207. [Google Scholar] [CrossRef]
- Fonseca, M.C.; Nascimento, C.S.; Borges, K.B. Theoretical investigation on functional monomer and solvent selection for molecular imprinting of tramadol. Chem. Phys. Lett. 2016, 645, 174–179. [Google Scholar] [CrossRef]
- Zhang, B.; Fan, X.; Zhao, D. Computer-aided design of molecularly imprinted polymers for simultaneous detection of clenbuterol and its metabolites. Polymers 2018, 11, 17. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; He, L.; Wang, Q.; Tang, Q.; Liu, F. Theoretical and experimental studies of a novel electrochemical sensor based on molecularly imprinted polymer and GQDs-PtNPs nanocomposite. Microchem. J. 2020, 158, 105196. [Google Scholar] [CrossRef]
- Huang, X.; Zhang, W.; Wu, Z.; Li, H.; Yang, C.; Ma, W.; Hui, A.; Zeng, Q.; Xiong, B.; Xian, Z. Computer simulation aided preparation of molecularly imprinted polymers for separation of bilobalide. J. Mol. Model. 2020, 26, 198. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Xiao, N.; Li, L.; Xie, X.; Li, Y. Theoretical insight into the interaction between chloramphenicol and functional monomer (methacrylic acid) in molecularly imprinted polymers. Int. J. Mol. Sci. 2020, 21, 4139. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yang, F.; Zhao, X.; Li, Y. Screening of functional monomers and solvents for the molecular imprinting of paclitaxel separation: A theoretical study. J. Mol. Model. 2020, 26, 26. [Google Scholar] [CrossRef]
- Silva, C.F.; Borges, K.B.; Nascimento, C.S. Computational study on acetamiprid-molecular imprinted polymer. J. Mol. Model. 2019, 25, 1–5. [Google Scholar] [CrossRef]
- Khodadadian, M.; Ahmadi, F. Computer-assisted design and synthesis of molecularly imprinted polymers for selective extraction of acetazolamide from human plasma prior to its voltammetric determination. Talanta 2010, 81, 1446–1453. [Google Scholar] [CrossRef]
- Wang, H.; Qian, D.; Xiao, X.; Gao, S.; Cheng, J.; He, B.; Liao, L.; Deng, J. A highly sensitive and selective sensor based on a graphene-coated carbon paste electrode modified with a computationally designed boron-embedded duplex molecularly imprinted hybrid membrane for the sensing of lamotrigine. Biosens. Bioelectron. 2017, 94, 663–670. [Google Scholar] [CrossRef]
- Gholivand, M.B.; Torkashvand, M.; Malekzadeh, G. Fabrication of an electrochemical sensor based on computationally designed molecularly imprinted polymers for determination of cyanazine in food samples. Anal. Chim. Acta 2012, 713, 36–44. [Google Scholar] [CrossRef]
- Yu, R.; Zhou, H.; Li, M.; Song, Q. Rational selection of the monomer for molecularly imprinted polymer preparation for selective and sensitive detection of 3-methylindole in water. J. Electroanal. Chem. 2019, 832, 129–136. [Google Scholar] [CrossRef]
- Marc, M.; Panuszko, A.; Namiesnik, J.; Wieczorek, P.P. Preparation and characterization of dummy-template molecularly imprinted polymers as potential sorbents for the recognition of selected polybrominated diphenyl ethers. Anal. Chim. Acta 2018, 1030, 77–95. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Lay, S.; Yu, H.; Shen, S. Synthesis and application of selective adsorbent for pirimicarb pesticides in aqueous media using allyl-β -cyclodextrin based binary functional monomers. J. Sci. Food Agric. 2018, 98, 2089–2097. [Google Scholar] [CrossRef]
- Nezhadali, A.; Mojarrab, M. Computational design and multivariate optimization of an electrochemical metoprolol sensor based on molecular imprinting in combination with carbon nanotubes. Anal. Chim. Acta 2016, 924, 86–98. [Google Scholar] [CrossRef]
- Gao, B.; He, X.-P.; Jiang, Y.; Wei, J.-T.; Suo, H.; Zhao, C. Computational simulation and preparation of fluorescent magnetic molecularly imprinted silica nanospheres for ciprofloxacin or norfloxacin sensing. J. Sep. Sci. 2014, 37, 3753–3759. [Google Scholar] [CrossRef] [PubMed]
- Tiu, B.D.B.; Pernites, R.B.; Tiu, S.B.; Advincula, R.C. Detection of aspartame via microsphere-patterned and molecularly imprinted polymer arrays. Colloids Surf. A Physicochem. Eng. Asp. 2016, 495, 149–158. [Google Scholar] [CrossRef] [Green Version]
- Vergara, A.V.; Pernites, R.B.; Pascua, S.; Binag, C.A.; Advincula, R.C. QCM sensing of a chemical nerve agent analog via electropolymerized molecularly imprinted polythiophene films. J. Polym. Sci. Part A Polym. Chem. 2012, 50, 675–685. [Google Scholar] [CrossRef]
- Nezhadali, A.; Shadmehri, R. Computer-aided sensor design and analysis of thiocarbohydrazide in biological matrices using electropolymerized-molecularly imprinted polypyrrole modified pencil graphite electrode. Sens. Actuators B Chem. 2013, 177, 871–878. [Google Scholar] [CrossRef]
- Khan, M.S.; Wate, P.S.; Krupadam, R.J. Combinatorial screening of polymer precursors for preparation of benzo[α] pyrene imprinted polymer: An ab initio computational approach. J. Mol. Model. 2012, 18, 1969–1981. [Google Scholar] [CrossRef] [PubMed]
- Prasad, B.B.; Rai, G. Study on monomer suitability toward the template in molecularly imprinted polymer: An ab initio approach. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2012, 88, 82–89. [Google Scholar] [CrossRef]
- Gholivand, M.B.; Khodadadian, M.; Ahmadi, F. Computer aided-molecular design and synthesis of a high selective molecularly imprinted polymer for solid-phase extraction of furosemide from human plasma. Anal. Chim. Acta 2010, 658, 225–232. [Google Scholar] [CrossRef]
- Ganjavi, F.; Ansari, M.; Kazemipour, M.; Zeidabadinejad, L. Computer-aided design and synthesis of a highly selective molecularly imprinted polymer for the extraction and determination of buprenorphine in biological fluids. J. Sep. Sci. 2017, 40, 3175–3182. [Google Scholar] [CrossRef]
- Azimi, A.; Javanbakht, M. Computational prediction and experimental selectivity coefficients for hydroxyzine and cetirizine molecularly imprinted polymer based potentiometric sensors. Anal. Chim. Acta 2014, 812, 184–190. [Google Scholar] [CrossRef]
- Hasanah, A.N.; Rahayu, D.; Pratiwi, R.; Rostinawati, T.; Megantara, S.; Saputri, F.A.; Puspanegara, K.H. Extraction of atenolol from spiked blood serum using a molecularly imprinted polymer sorbent obtained by precipitation polymerization. Heliyon 2019, 5, e01533. [Google Scholar] [CrossRef] [Green Version]
- Hasanah, A.N.; Soni, D.; Pratiwi, R.; Rahayu, D.; Megantara, S.; Mutakin. Synthesis of diazepam-imprinted polymers with two functional monomers in chloroform using a bulk polymerization method. J. Chem. 2020, 2020, 7282415. [Google Scholar] [CrossRef]
- Tabandeh, M.; Ghassamipour, S.; Aqababa, H.; Tabatabaei, M.; Hasheminejad, M. Computational design and synthesis of molecular imprinted polymers for selective extraction of allopurinol from human plasma. J. Chromatogr. B 2012, 898, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, F.; Rezaei, H.; Tahvilian, R. Computational-aided design of molecularly imprinted polymer for selective extraction of methadone from plasma and saliva and determination by gas chromatography. J. Chromatogr. A 2012, 1270, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Aqababa, H.; Tabandeh, M.; Tabatabaei, M.; Hasheminejad, M.; Emadi, M. Computer-assisted design and synthesis of a highly selective smart adsorbent for extraction of clonazepam from human serum. Mater. Sci. Eng. C 2013, 33, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Salajegheh, M.; Ansari, M.; Foroghi, M.M.; Kazemipour, M. Computational design as a green approach for facile preparation of molecularly imprinted polyarginine-sodium alginate-multiwalled carbon nanotubes composite film on glassy carbon electrode for theophylline sensing. J. Pharm. Biomed. Anal. 2019, 162, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Hussain, S.; Wong, A.; Foguel, M.V.; Moreira Gonçalves, L.; Pividori Gurgo, M.I.; Taboada Sotomayor, M.D.P. Synthesis and characterization of magnetic-molecularly imprinted polymers for the HPLC-UV analysis of ametryn. React. Funct. Polym. 2018, 122, 175–182. [Google Scholar] [CrossRef] [Green Version]
- Nashar, R.M.E.; Ghani, N.T.A.; Gohary, N.A.E.; Barhoum, A.; Madbouly, A. Molecularly imprinted polymers based biomimetic sensors for mosapride citrate detection in biological fluids. Mater. Sci. Eng. C 2017, 76, 123–129. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Gu, X.; Shi, L.; Hong, J.; Zhang, H.; Gao, Y.; Du, S.; Chen, L. Molecularly imprinted polymers based on SBA-15 for selective solid-phase extraction of baicalein from plasma samples. Anal. Bioanal. Chem. 2015, 407, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Lipkowitz, K.B.; Peterson, M.A. Molecular mechanics in organic synthesis. Chem. Rev. 1993, 93, 2463–2486. [Google Scholar] [CrossRef]
- Karlsson, B.C.G.; O’Mahony, J.; Karlsson, J.G.; Bengtsson, H.; Eriksson, L.A.; Nicholls, I.A. Structure and dynamics of monomer–template Complexation: An Explanation for Molecularly Imprinted Polymer Recognition Site Heterogeneity. J. Am. Chem. Soc. 2009, 131, 13297–13304. [Google Scholar] [CrossRef] [PubMed]
- Alder, B.J.; Wainwright, T.E. Phase transition for a hard sphere system. J. Chem. Phys. 1957, 27, 1208–1209. [Google Scholar] [CrossRef] [Green Version]
- Wormer, P.E.S.; Avoird, A.V.D. Chapter 37—Forty years of ab initio calculations on intermolecular forces. Theory Appl. Comput. Chem. 2005, 1047–1077. [Google Scholar]
- Seminario, J.M. Chapter 6—Ab initio and DFT for the strength of classical molecular dynamics. Theor. Comput. Chem. 1999, 7, 187–229. [Google Scholar]
- Boeyens, J.C.A.; Comba, P. ChemInform abstract: Molecular mechanics: Theoretical basis, rules, scope and limits. ChemInform 2001, 32, 3–10. [Google Scholar] [CrossRef]
- Kazemi, S.; Daryani, A.S.; Abdouss, M.; Shariatinia, Z. DFT computations on the hydrogen bonding interactions between methacrylic acid-trimethylolpropane trimethacrylate copolymers and letrozole as drug delivery systems. J. Theor. Comput. Chem. 2016, 15, 1–20. [Google Scholar] [CrossRef]
- Huang, Y.; Zhu, Q. Computational modeling and theoretical calculations on the interactions between spermidine and functional monomer (methacrylic acid) in a molecularly imprinted polymer. J. Chem. 2015, 2015, 216983. [Google Scholar] [CrossRef]
- Li, L.; Chen, L.; Zhang, H.; Yang, Y.; Liu, X.; Chen, Y. Temperature and magnetism bi-responsive molecularly imprinted polymers: Preparation, adsorption mechanism and properties as drug delivery system for sustained release of 5-fluorouracil. Mater. Sci. Eng. C 2016, 61, 158–168. [Google Scholar] [CrossRef] [Green Version]
- Cohen, A.J.; Mori-Sánchez, P.; Yang, W. Challenges for density functional theory. Chem. Rev. 2012, 112, 289–320. [Google Scholar] [CrossRef] [PubMed]
- Kohn, W.; Sham, L.J. Self-Consistent Equations Including Exchange and Correlation Effects. Phys. Rev. 1965, 140, A1133–A1138. [Google Scholar] [CrossRef] [Green Version]
- Dong, W.; Yan, M.; Zhang, M.; Liu, Z.; Li, Y. A computational and experimental investigation of the interaction between the template molecule and the functional monomer used in the molecularly imprinted polymer. Anal. Chim. Acta 2005, 542, 186–192. [Google Scholar] [CrossRef]
- Wu, L.; Sun, B.; Li, Y.; Chang, W. Study properties of molecular imprinting polymer using a computational approach. Analyst 2003, 128, 944. [Google Scholar] [CrossRef]
- Nicholls, I.A.; Andersson, H.S.; Charlton, C.; Henschel, H.; Karlsson, B.C.G.; Karlsson, J.G.; O’Mahony, J.; Rosengren, A.M.; Rosengren, K.J.; Wikman, S. Theoretical and computational strategies for rational molecularly imprinted polymer design. Biosens. Bioelectron. 2009, 25, 543–552. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chen, L.; Zhang, H.; Yang, Y.; Liu, X. Recognition of 5-fluorouracil by thermosensitive magnetic surface molecularly imprinted microspheres designed using a computational approach. J. Appl. Polym. Sci. 2017, 134, 45468. [Google Scholar] [CrossRef]
- Dong, W.; Yan, M.; Liu, Z.; Wu, G.; Li, Y. Effects of solvents on the adsorption selectivity of molecularly imprinted polymers: Molecular simulation and experimental validation. Sep. Purif. Technol. 2007, 53, 183–188. [Google Scholar] [CrossRef]
- Douhaya, Y.V.; Barkaline, V.V.; Tsakalof, A. Computer-simulation-based selection of optimal monomer for imprinting of tri-O-acetyl adenosine in a polymer matrix: Calculations for benzene solution. J. Mol. Model. 2016, 22, 157. [Google Scholar] [CrossRef]
- Qader, B.; Baron, M.; Hussain, I.; Gonzalez-Rodriguez, J. Electrochemical determination of 2-isopropoxyphenol in glassy carbon and molecularly imprinted poly-pyrrole electrodes. J. Electroanal. Chem. 2018, 821, 16–21. [Google Scholar] [CrossRef]
- Qader, B.; Baron, M.; Hussain, I.; Sevilla, J.M.; Johnson, R.P.; Gonzalez-Rodriguez, J. Electrochemical determination of disulfoton using a molecularly imprinted poly-phenol polymer. Electrochim. Acta 2019, 295, 333–339. [Google Scholar] [CrossRef]
- Peng, M.; Li, H.; Long, R.; Shi, S.; Zhou, H.; Yang, S. Magnetic porous molecularly imprinted polymers based on surface precipitation polymerization and mesoporous SiO2 layer as sacrificial support for efficient and selective extraction and determination of chlorogenic acid in duzhong brick Tea. Molecules 2018, 23, 1554. [Google Scholar]
- Ayankojo, A.G.; Reut, J.; Boroznjak, R.; Öpik, A.; Syritski, V. Molecularly imprinted poly(meta-phenylenediamine) based QCM sensor for detecting Amoxicillin. Sens. Actuators B Chem. 2018, 258, 766–774. [Google Scholar] [CrossRef]
- Li, H.; He, H.; Huang, J.; Wang, C.-Z.; Gu, X.; Gao, Y.; Zhang, H.; Du, S.; Chen, L.; Yuan, C.-S. A novel molecularly imprinted method with computational simulation for the affinity isolation and knockout of baicalein from Scutellaria baicalensis. Biomed. Chromatogr. 2016, 30, 117–125. [Google Scholar] [CrossRef]
- Pereira, T.F.D.; Da Silva, A.T.M.; Borges, K.B.; Nascimento, C.S. Carvedilol-imprinted polymer: Rational design and selectivity studies. J. Mol. Struct. 2019, 1177, 101–106. [Google Scholar] [CrossRef]
- Kowalska, A.; Stobiecka, A.; Wysocki, S. A computational investigation of the interactions between harmane and the functional monomers commonly used in molecular imprinting. J. Mol. Struct. 2009, 901, 88–95. [Google Scholar] [CrossRef]
- Rebelo, P.; Pacheco, J.G.; Voroshylova, I.V.; Melo, A.; Cordeiro, M.N.D.S.; Delerue-Matos, C. Rational development of molecular imprinted carbon paste electrode for Furazolidone detection: Theoretical and experimental approach. Sens. Actuators B Chem. 2021, 329, 129112. [Google Scholar] [CrossRef]
- Wang, D.; Yang, Y.; Xu, Z.; Liu, Y.; Liu, Z.; Lin, T.; Chen, X.; Liu, H. Molecular simulation-aided preparation of molecularly imprinted polymeric solid-phase microextraction coatings for kojic acid detection in wheat starch and flour samples. Food Anal. Methods 2021, 1–12. [Google Scholar] [CrossRef]
- Madikizela, L.M.; Mdluli, P.S.; Chimuka, L. Experimental and theoretical study of molecular interactions between 2-vinyl pyridine and acidic pharmaceuticals used as multi-template molecules in molecularly imprinted polymer. React. Funct. Polym. 2016, 103, 33–43. [Google Scholar] [CrossRef]
- Wu, L.; Zhu, K.; Zhao, M.; Li, Y. Theoretical and experimental study of nicotinamide molecularly imprinted polymers with different porogens. Anal. Chim. Acta 2005, 549, 39–44. [Google Scholar] [CrossRef]
- Ahmadi, F.; Sadeghi, T.; Ataie, Z.; Rahimi-Nasrabadi, M.; Eslami, N. Computational design of a selective molecular imprinted polymer for extraction of pseudoephedrine from plasma and determination by HPLC. Anal. Chem. Lett. 2017, 7, 295–310. [Google Scholar] [CrossRef]
- Han, Y.; Gu, L.; Zhang, M.; Li, Z.; Yang, W.; Tang, X.; Xie, G. Computer-aided design of molecularly imprinted polymers for recognition of atrazine. Comput. Theor. Chem. 2017, 1121, 29–34. [Google Scholar] [CrossRef]
- Baggiani, C.; Anfossi, L.; Baravalle, P.; Giovannoli, C.; Tozzi, C. Selectivity features of molecularly imprinted polymers recognising the carbamate group. Anal. Chim. Acta 2005, 531, 199–207. [Google Scholar] [CrossRef]
- Piletska, E.V.; Turner, N.W.; Turner, A.P.F.; Piletsky, S.A. Controlled release of the herbicide simazine from computationally designed molecularly imprinted polymers. J. Control. Release 2005, 108, 132–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sergeyeva, T.A.; Piletska, O.V.; Piletsky, S.A.; Sergeeva, L.M.; Brovko, O.O.; El’Ska, G.V. Data on the structure and recognition properties of the template-selective binding sites in semi-IPN-based molecularly imprinted polymer membranes. Mater. Sci. Eng. C 2008, 28, 1472–1479. [Google Scholar] [CrossRef]
- Altintas, Z.; France, B.; Ortiz, J.O.; Tothill, I.E. Computationally modelled receptors for drug monitoring using an optical based biomimetic SPR sensor. Sens. Actuators B Chem. 2016, 224, 726–737. [Google Scholar] [CrossRef]
- Altintas, Z.; Abdin, M.J.; Tothill, A.M.; Karim, K.; Tothill, I.E. Ultrasensitive detection of endotoxins using computationally designed nanoMIPs. Anal. Chim. Acta 2016, 935, 239–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sergeyeva, T.A.; Piletsky, S.A.; Brovko, A.A.; Slinchenko, E.A.; Sergeeva, L.M.; El’Skaya, A.V. Selective recognition of atrazine by molecularly imprinted polymer membranes. Development of conductometric sensor for herbicides detection. Anal. Chim. Acta 1999, 392, 105–111. [Google Scholar] [CrossRef]
- Liang, S.; Wan, J.; Zhu, J.; Cao, X. Effects of porogens on the morphology and enantioselectivity of core–shell molecularly imprinted polymers with ursodeoxycholic acid. Sep. Purif. Technol. 2010, 72, 208–216. [Google Scholar] [CrossRef]
- Viveiros, R.; Karim, K.; Piletsky, S.A.; Heggie, W.; Casimiro, T. Development of a molecularly imprinted polymer for a pharmaceutical impurity in supercritical CO2: Rational design using computational approach. J. Clean. Prod. 2017, 168, 1025–1031. [Google Scholar] [CrossRef]
- Feng, F.; Zheng, J.W.; Qin, P.; Han, T.; Zhao, D.Y. A novel quartz crystal microbalance sensor array based on molecular imprinted polymers for simultaneous detection of clenbuterol and its metabolites. Talanta 2017, 167, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Sobiech, M.; Żołek, T.; Luliński, P.; Maciejewska, D. Separation of octopamine racemate on (R, S)-2-amino-1-phenylethanol imprinted polymer–Experimental and computational studies. Talanta 2016, 146, 556–567. [Google Scholar] [CrossRef]
- Erracina, J.J.; Sharfstein, S.T.; Bergkvist, M. In silico characterization of enantioselective molecularly imprinted binding sites. J. Mol. Recognit. 2018, 31, e2612. [Google Scholar]
- Nascimento, T.A.; Silva, C.F.; Oliveira, H.L.D.; Da Silva, R.C.S.; Nascimento, C.S.; Borges, K.B. Magnetic molecularly imprinted conducting polymer for determination of praziquantel enantiomers in milk. Analyst 2020, 145, 4245–4253. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, R.A.; Dinali, L.A.F.; Silva, C.F.; De Oliveira, H.L.; Da Silva, A.T.M.; Nascimento, C.S.; Borges, K.B. Microextraction by packed molecularly imprinted polymer followed by ultra-high performance liquid chromatography for determination of fipronil and fluazuron residues in drinking water and veterinary clinic wastewater. Microchem. J. 2021, 168, 106405. [Google Scholar] [CrossRef]
- Lu, J.; Qin, Y.; Wu, Y.; Chen, M.; Sun, C.; Han, Z.; Yan, Y.; Li, C.; Yan, Y. Mimetic-core-shell design on molecularly imprinted membranes providing an antifouling and high-selective surface. Chem. Eng. J. 2021, 417, 128085. [Google Scholar] [CrossRef]
- Zhang, Y.; Tan, X.; Liu, X.; Li, C.; Zeng, S.; Wang, H.; Zhang, S. Fabrication of multilayered molecularly imprinted membrane for selective recognition and separation of artemisinin. ACS Sustain. Chem. Eng. 2019, 7, 3127–3137. [Google Scholar] [CrossRef]
- Wang, J.; Guo, R.; Chen, J.; Zhang, Q.; Liang, X. Phenylurea herbicides-selective polymer prepared by molecular imprinting using N-(4-isopropylphenyl)-N′-butyleneurea as dummy template. Anal. Chim. Acta 2005, 540, 307–315. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, D.; Du, F.; Zheng, W.; Liu, Z.; Xu, Z.; Hu, X.; Liu, H. Dummy-template molecularly imprinted micro-solid-phase extraction coupled with high-performance liquid chromatography for bisphenol A determination in environmental water samples. Microchem. J. 2019, 145, 337–344. [Google Scholar] [CrossRef]




| Simulation Method | Template | Force Field/Method | Software | MIPs Design |
|---|---|---|---|---|
| Molecular mechanics (MM) | Myoglobin [29] | OPLS3 | Prime | Screening functional monomers |
| Morphine [30] | CHARMM and MMFF94 | Discovery Studio | Template-monomer ratio | |
| Metolachlor deschloro [31], metsulfuron-methyl [32] | AMBER MM | SPSS Statistics | Screening functional monomers/template-monomer ratio | |
| Norfloxacin [33] | MMFF94X | Discovery Studio | Screening functional monomers/template-monomer ratio | |
| Molecular dynamics (MD) | Curcumin [34], fenthion [35], N-3-oxo-dodecanoyl-L-homoserine lactone [36], methidathion [37], endotoxins [38], phosmet insecticide [39], cocaine [40], methyl parathion [41], aflatoxin B1 [42] | Tripos | SYBYL | Screening functional monomers/template-monomer ratio |
| Bisphenol A [43], carbamazepine [44], phthalates [45], norfloxacin [46], sulfamethoxazole [47] | COMPASS | Materials Studio/accelrys.com | Screening functional monomers/template-monomer ratio | |
| Thiamethoxam [48] | AMBER | Gaussian | Template-monomer ratio and solvent | |
| Rhodamine B [49] | GROMOS | GROMACS | Template-monomer ratio and solvent | |
| Quantum mechanics (QM) | Vancomycin [50], primaquine [51], tramadol [52], thiamethoxam [48], clenbuterol [53], sulfadimidine [54], bilobalide [55], chloramphenicol [56], paclitaxel [57], acetamiprid [58], acetazolamide [59], lamotrigine [60], cyanazine [61], 3-methylindole [62], polybrominated diphenyl ethers [63], pirimicarb [64], metoprolol [65], ciprofloxacin or norfloxacin [66] | DFT | Gaussian | Screening functional monomers/template-monomer ratio/solvent |
| Aspartame [67], pinacolyl methylphosphonate [68], metolachlor deschloro [31], metsulfuron-methyl [32], thiocarbohydrazide [69] | Semiempirical method | Spartan/SPSS Statistics | Screening functional monomers/template-monomer ratio | |
| Benzo[a]pyrene [70], tryptophan [71], furosemide [72], buprenorphine [73], hydroxyzine and cetirizine [74], atenolol [75], diazepam [76], metolachlor deschloro [31], metsulfuron-methyl [32], allopurinol [77], methadone [78], clonazepam [79], theophylline [80], ametryn [81], mosapride citrate [82], baicalein [83], | Ab initio | HyperChem/Gaussian/AutoDockTools/SPSS Statistics | Screening functional monomers/template-monomer ratio |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Xu, Z.; Wang, D.; Yang, Y.; Duan, Y.; Ma, L.; Lin, T.; Liu, H. A Review on Molecularly Imprinted Polymers Preparation by Computational Simulation-Aided Methods. Polymers 2021, 13, 2657. https://doi.org/10.3390/polym13162657
Liu Z, Xu Z, Wang D, Yang Y, Duan Y, Ma L, Lin T, Liu H. A Review on Molecularly Imprinted Polymers Preparation by Computational Simulation-Aided Methods. Polymers. 2021; 13(16):2657. https://doi.org/10.3390/polym13162657
Chicago/Turabian StyleLiu, Zhimin, Zhigang Xu, Dan Wang, Yuming Yang, Yunli Duan, Liping Ma, Tao Lin, and Hongcheng Liu. 2021. "A Review on Molecularly Imprinted Polymers Preparation by Computational Simulation-Aided Methods" Polymers 13, no. 16: 2657. https://doi.org/10.3390/polym13162657