Highly Selective Enzymatic Recovery of Building Blocks from Wool-Cotton-Polyester Textile Waste Blends
Abstract
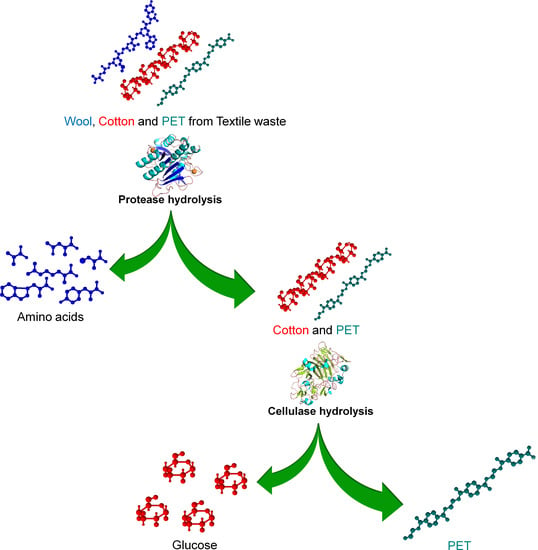
:1. Introduction
2. Materials and Methods
2.1. Chemicals, Substrates and Enzymes
2.2. Enzymatic Hydrolysis
2.3. Protein Hydrolysate Characterization
2.3.1. Protease Activity and Thermal Stability Assay
2.3.2. Chemical Analysis
2.3.3. Molecular Weight Distribution of Keratin Hydrolysis
2.3.4. Amino Acid Determination
2.4. Cellulose Hydrolysis
2.4.1. Total Cellulose Assay
2.4.2. Quantification of Glucose via HPLC
2.4.3. Bio-Ethanol Production
2.5. Poly(Ethylene Terephthalate) Characterization
3. Results and Discussion
3.1. Step-Wise Enzymatic Extraction of Textile Building Blocks
3.2. Cellulose-Based Material Hydrolysis and Ethanol Production
3.3. Characterization of the Recovered Poly(Ethylene Terephthalate) (PET)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wang, Y. Carpet Fiber Recycling Technologies. Ecotextiles 2007, 26–32. [Google Scholar] [CrossRef]
- Zamani, B. Towards Understanding Sustainable Textile Waste Management: Environmental Impacts and Social Indicators. Ph.D. Thesis, Chalmers University, Gothenburg, Sweden, 2014; pp. 1–52. [Google Scholar]
- Payne, A. 6-Open- and closed-loop recycling of textile and apparel products. In Handbook of Life Cycle Assessment (LCA) of Textiles and Clothing; Elsevier Ltd.: Amsterdam, The Netherlands, 2015; pp. 103–123. [Google Scholar] [CrossRef]
- Oakdene Hollins. Chemical Recycling—A Solution for Europe’s Waste Textile Mountain? Available online: https://www.oakdenehollins.com/news-insights/2016/12/16/chemical-recycling-a-solution-for-europes-waste-textile-mountain (accessed on 2 September 2018).
- Gentil, E.; Clavreul, J.; Christensen, T.H. Global warming factor of municipal solid waste management in Europe. Waste Manag. Res. 2009, 27, 850–860. [Google Scholar] [CrossRef] [PubMed]
- Hawley, J.M. Textile Recycling. In Handbook of Recycling: State-of-the-Art for Practitioners, Analysts, and Scientists; Elsevier Ltd.: Amsterdam, The Netherlands, 2014; pp. 211–217. [Google Scholar] [CrossRef]
- Cucchiella, F.; D’Adamo, I.; Gastaldi, M. Sustainable management of waste-to-energy facilities. Renew. Sustain. Energy Rev. 2014, 33, 719–728. [Google Scholar] [CrossRef]
- Cole, C.; Osmani, M.; Quddus, M.; Wheatley, A.; Kay, K. Towards a zero waste strategy for an English local authority. Resour. Conserv. Recycl. 2014, 89, 64–75. [Google Scholar] [CrossRef]
- Lu, J.; Hamouda, H. Current Status of Fiber Waste Recycling and Its Future. Adv. Mater. Res. 2014, 878, 122–131. [Google Scholar] [CrossRef]
- Towards a 4th Industrial Revolution of Textiles and Clothing. Available online: https://www.google.com/url?sa=t&rct=j&q=&esrc=s&source=web&cd=1&ved=2ahUKEwjcl_CalfHdAhWTQN4KHR2hBNUQFjAAegQICRAC&url=http%3A%2F%2Fstatic1.1.sqspcdn.com%2Fstatic%2Ff%2F834046%2F27293567%2F1476721468317%2FTextileETP_SIRA_public%2Bversion.pdf%3Ftoken%3D3XnfEgvscghojs5KRKgx8vSWa5M%253D&usg=AOvVaw22adV1ZvagZ4ELQQ6px9Pd (accessed on 2 September 2018).
- Van Der Velden, N.M.; Patel, M.K.; Vogtländer, J.G. LCA benchmarking study on textiles made of cotton, polyester, nylon, acryl, or elastane. Int. J. Life Cycle Assess. 2014, 19, 331–356. [Google Scholar] [CrossRef]
- Araújo, R.; Casal, M.; Cavaco-Paulo, A. Application of enzymes for textile fibres processing. Biocatal. Biotransform. 2008, 26, 332–349. [Google Scholar] [CrossRef] [Green Version]
- Chavan, R. Environmental Sustainability through Textile Recycling. J. Text. Sci. Eng. 2014, s2, 1–5. [Google Scholar] [CrossRef]
- Pellis, A.; Cantone, S.; Ebert, C.; Gardossi, L. Evolving biocatalysis to meet bioeconomy challenges and opportunities. N. Biotechnol. 2018, 40, 154–169. [Google Scholar] [CrossRef] [PubMed]
- Guebitz, G.M.; Cavaco-Paulo, A. Enzymes go big: Surface hydrolysis and functionalisation of synthetic polymers. Trends Biotechnol. 2008, 26, 32–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buschle-Diller, G.; Zeronian, S.H.; Pan, N.; Yoon, M.Y. Enzymatic Hydrolysis of Cotton, Linen, Ramie, and Viscose Rayon Fabrics. Text. Res. J. 1994, 64, 270–279. [Google Scholar] [CrossRef]
- Eslahi, N.; Dadashian, F.; Nejad, N.H. An investigation on keratin extraction from wool and feather waste by enzymatic hydrolysis. Prep. Biochem. Biotechnol. 2013, 43, 624–648. [Google Scholar] [CrossRef] [PubMed]
- Gamerith, C.; Zartl, B.; Pellis, A.; Guillamot, F.; Marty, A.; Acero, E.H.; Guebitz, G.M. Enzymatic recovery of polyester building blocks from polymer blends. Process Biochem. 2017, 59, 58–64. [Google Scholar] [CrossRef]
- Vecchiato, S.; Skopek, L.; Jankova, S.; Pellis, A.; Ipsmiller, W.; Aldrian, A.; Mueller, B.; Acero, E.H.; Guebitz, G.M. Enzymatic Recycling of High-Value Phosphor Flame-Retardant Pigment and Glucose from Rayon Fibers. ACS Sustain. Chem. Eng. 2018, 6, 2386–2394. [Google Scholar] [CrossRef]
- Weinberger, S.; Haernvall, K.; Scaini, D.; Ghazaryan, G.; Zumstein, M.T.; Sander, M.; Pellis, A.; Guebitz, G.M. Enzymatic surface hydrolysis of poly(ethylene furanoate) thin films of various crystallinities. Green Chem. 2017, 19, 5381–5384. [Google Scholar] [CrossRef]
- Eslahi, N.; Dadashian, F.; Nejad, N.H. Optimization of enzymatic hydrolysis of wool fibers for nanoparticles production using response surface methodology. Adv. Powder Technol. 2013, 24, 416–426. [Google Scholar] [CrossRef]
- Novozymes Cellic® CTec3–Secure Your Plant’s Lowest Total Cost. Available online: http://s3.amazonaws.com/zanran_storage/bioenergy.novozymes.com/ContentPages/2546502386.pdf (accessed on 2 September 2018).
- Brock, F.M.; Forsberg, C.W.; Buchanan-Smith, J.G. Proteolytic activity of rumen microorganisms and effects of proteinase inhibitors. Appl. Environ. Microbiol. 1982, 44, 561–569. [Google Scholar]
- Bhavsar, P.; Zoccola, M.; Patrucco, A.; Montarsolo, A.; Mossotti, R.; Rovero, G.; Giansetti, M.; Tonin, C. Superheated Water Hydrolysis of Waste Wool in a Semi-Industrial Reactor to Obtain Nitrogen Fertilizers. ACS Sustain. Chem. Eng. 2016, 4, 6722–6731. [Google Scholar] [CrossRef]
- Ghose, T.K. Measurement of cellulase activities. Pure Appl. Chem. 1987, 59, 257–268. [Google Scholar] [CrossRef] [Green Version]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Plowman, J.E. Proteomic database of wool components. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2003, 787, 63–76. [Google Scholar] [CrossRef]
- Röper, K.; Föhles, J.; Klostermeyer, H. Complete Enzymatic Hydrolysis of Wool and Its Morphological Components. Methods Enzymol. 1984, 106, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Betzel, C.; Klupsch, S.; Papendorf, G.; Hastrup, S.; Branner, S.; Wilson, K.S. Crystal structure of the alkaline proteinase SavinaseTM from Bacillus Ientus at 1.4 Å resolution. J. Mol. Biol. 1992, 223, 427–445. [Google Scholar] [CrossRef]
- Nustorova, M.; Braikova, D.; Gousterova, A.; Vasileva-Tonkova, E.; Nedkov, P. Chemical, microbiological and plant analysis of soil fertilized with alkaline hydrolysate of sheep’s wool waste. World J. Microbiol. Biotechnol. 2006, 22, 383–390. [Google Scholar] [CrossRef]
- Bhavsar, P.; Zoccola, M.; Patrucco, A.; Montarsolo, A.; Rovero, G.; Tonin, C. Comparative study on the effects of superheated water and high temperature alkaline hydrolysis on wool keratin. Text. Res. J. 2017, 87, 1696–1705. [Google Scholar] [CrossRef]
- Valverde, I.C.; Castilla, L.H.; Nuñez, D.F.; Rodriguez-Senín, E.; De La Mano Ferreira, R. Development of new insulation panels based on textile recycled fibers. Waste Biomass Valorization 2013, 4, 139–146. [Google Scholar] [CrossRef]
- Moubarik, A.; Pizzi, A.; Allal, A.; Charrier, F.; Khoukh, A.; Charrier, B. Cornstarch-mimosa tannin-urea formaldehyde resins as adhesives in the particleboard production. Starch 2010, 62, 131–138. [Google Scholar] [CrossRef]
- Binici, H.; Eken, M.; Kara, M.; Dolaz, M. An environment-friendly thermal insulation material from sunflower stalk, textile waste and stubble fibers. Proc. Int. Conf. Renew. Energy Res. Appl. ICRERA 2013, 51, 833–846. [Google Scholar] [CrossRef]
- Dimarogona, M.; Topakas, E.; Christakopoulos, P. Cellulose Degradation by Oxidative Enzymes. Comput. Struct. Biotechnol. J. 2012, 2, e201209015. [Google Scholar] [CrossRef] [PubMed]
- López-Linares, J.C.; Romero, I.; Cara, C.; Ruiz, E.; Moya, M.; Castro, E. Bioethanol production from rapeseed straw at high solids loading with different process configurations. Fuel 2014, 122, 112–118. [Google Scholar] [CrossRef]
- Pellis, A.; Herrero Acero, E.; Ferrario, V.; Ribitsch, D.; Guebitz, G.M.; Gardossi, L. The Closure of the Cycle: Enzymatic Synthesis and Functionalization of Bio-Based Polyesters. Trends Biotechnol. 2016, 34, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Ortner, A.; Pellis, A.; Gamerith, C.; Orcal Yebra, A.; Scaini, D.; Kaluzna, I.; Mink, D.; de Wildeman, S.; Herrero Acero, E.; Guebitzab, G.M. Superhydrophobic functionalization of cutinase activated poly(lactic acid) surfaces. Green Chem. 2017, 19, 816–822. [Google Scholar] [CrossRef]
- Pellis, A.; Silvestrini, L.; Scaini, D.; Coburn, J.M.; Gardossi, L.; Kaplan, D.L.; Herrero Acero, E.; Guebitz, G.M. Enzyme-catalyzed functionalization of poly(l-lactic acid) for drug delivery applications. Process Biochem. 2017, 59, 77–83. [Google Scholar] [CrossRef]
- Quartinello, F.; Vajnhandl, S.; Volmajer Valh, J.; Farmer, T.J.; Vončina, B.; Lobnik, A.; Herrero Acero, E.; Pellis, A.; Guebitz, G.M. Synergistic chemo-enzymatic hydrolysis of poly(ethylene terephthalate) from textile waste. Microb. Biotechnol. 2017, 10, 1376–1383. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| Sample | Cotton (%) | Wool (%) | PET (%) |
|---|---|---|---|
| CWP_90/5/5 | 90 | 5 | 5 |
| CWP_80/5/10 | 80 | 5 | 15 |
| CWP_80/10/10 | 80 | 10 | 10 |
| CWP_70/10/20 | 70 | 10 | 20 |
| CWP_60/10/30 | 60 | 10 | 30 |
| CWP_50/10/40 | 50 | 10 | 40 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quartinello, F.; Vecchiato, S.; Weinberger, S.; Kremenser, K.; Skopek, L.; Pellis, A.; Guebitz, G.M. Highly Selective Enzymatic Recovery of Building Blocks from Wool-Cotton-Polyester Textile Waste Blends. Polymers 2018, 10, 1107. https://doi.org/10.3390/polym10101107
Quartinello F, Vecchiato S, Weinberger S, Kremenser K, Skopek L, Pellis A, Guebitz GM. Highly Selective Enzymatic Recovery of Building Blocks from Wool-Cotton-Polyester Textile Waste Blends. Polymers. 2018; 10(10):1107. https://doi.org/10.3390/polym10101107
Chicago/Turabian StyleQuartinello, Felice, Sara Vecchiato, Simone Weinberger, Klemens Kremenser, Lukas Skopek, Alessandro Pellis, and Georg M. Guebitz. 2018. "Highly Selective Enzymatic Recovery of Building Blocks from Wool-Cotton-Polyester Textile Waste Blends" Polymers 10, no. 10: 1107. https://doi.org/10.3390/polym10101107