Genome-Wide Identification of the YABBY Gene Family in Seven Species of Magnoliids and Expression Analysis in Litsea
Abstract
:1. Introduction
2. Results
2.1. Isolation and Sequence Analysis of YABBY Genes from Magnoliids
2.2. Congruence of YABBY and Zinc-Finger Domains
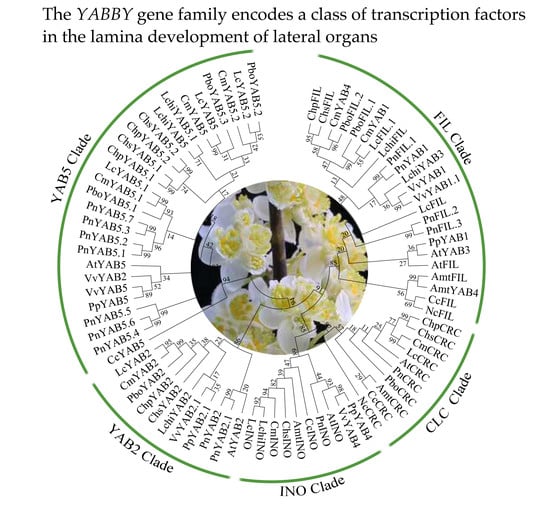
2.3. Phylogenetic Relationship and Gene Structure of YABBY Genes in Magnoliids
2.4. Gene Structure Analysis of YABBY Genes in Magnoliids
2.5. Gene Expression Analysis in Litsea
3. Discussion
4. Materials and Methods
4.1. Identification of the YABBY Gene Family in Magnoliids
4.2. Conserved Domains and Motifs in YABBY Proteins
4.3. Gene and Protein Structures Analysis
4.4. Phylogenetic Analyses of YABBY Gene Family in Magnoliids
4.5. Transcriptome Data Analysis and Gene Expression Heatmap
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bartholmes, C.; Hidalgo, O.; Gleissberg, S. Evolution of the YABBY gene family with emphasis on the basal eudicot Eschscholzia californica (Papaveraceae). Plant Biol. 2012, 14, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Finet, C.; Floyd, S.K.; Conway, S.J.; Zhong, B.; Scutt, C.P.; Bowman, J.L. Evolution of the YABBY gene family in seed plants. Evol. Dev. 2016, 18. [Google Scholar] [CrossRef] [PubMed]
- Filyushin, M.A.; Slugina, M.A.; Shchennikova, A.V.; Kochieva, E.Z. YABBY3-orthologous genes in wild tomato species: Structure, variability, and expression. Acta Nat. 2017, 9. [Google Scholar] [CrossRef] [Green Version]
- Kumaran, M.K.; Bowman, J.L.; Sundaresan, V. YABBY polarity genes mediate the repression of KNOX homeobox genes in Arab. Plant Cell 2002, 14, 2761–2770. [Google Scholar] [CrossRef]
- Floyd, S.K.; Bowman, J.L. Distinct developmental mechanisms reflect the independent origins of leaves in vascular plants. Curr. Biol. 2006, 16, 1911–1917. [Google Scholar] [CrossRef] [Green Version]
- Bowman, J.L.; Smyth, D.R. CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 1999, 126, 2387–2396. [Google Scholar] [CrossRef]
- Zhang, L.; Yang, Z.; Peng, Z.; Yu, Y.; Sun, Q. Molecular cloning, characterization, and expression analysis of DROOPING LEAF gene in the Pistillody Line of Common Wheat. Mod. Appl. Sci. 2015, 9, 44. [Google Scholar] [CrossRef]
- Meister, R.J.; Oldenhof, H.; Bowman, J.L.; Gasser, C.S. Multiple protein regions contribute to differential activities of YABBY proteins in reproductive development. Plant Physiol. 2005, 137, 651–662. [Google Scholar] [CrossRef] [Green Version]
- Sarojam, R.; Sappl, P.G.; Goldshmidt, A.; Efroni, I.; Floyd, S.K.; Eshed, Y.; Bowmana, J.L. Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell 2010, 22, 2113–2130. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Gong, Q.; Wang, L.; Jin, Y.; Xi, J.; Li, Z.; Qin, W.; Yang, Z.; Lu, L.; Chen, Q.; et al. Genome-wide study of YABBY genes in upland cotton and their expression patterns under different stresses. Front. Genet. 2018, 9, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Soundararajan, P.; Won, S.Y.; Park, D.S.; Lee, Y.H.; Sun Kim, J. Comparative analysis of the YABBY gene family of Bienertia sinuspersici, a single-cell c4 plant. Plants 2019, 8, 536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegfried, K.R.; Eshed, Y.; Baum, S.F.; Otsuga, D.; Drews, G.N.; Bowman, J.L. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 1999, 126, 4117–4128. [Google Scholar] [CrossRef] [PubMed]
- Pfannebecker, K.C.; Lange, M.; Rupp, O.; Becker, A.; Purugganan, M. Seed plant-specific gene lineages involved in carpel development. Mol. Biol. Evol. 2017, 34, 925–942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filyushin, M.A.; Slugin, M.A.; Dzhos, E.A.; Kochieva, E.Z.; Shchennikova, A.V. Coexpression of YABBY1 and YABBY3 genes in lateral organs of tomato species (Solanum, Section Lycopersicon). Dokl. Biochem. Biophys. 2018, 478, 50–54. [Google Scholar] [CrossRef] [PubMed]
- De Almeida, A.M.R.; Yockteng, R.; Schnable, J.; Alvarez-Buylla, E.R.; Freeling, M.; Specht, C.D. Co-option of the polarity gene network shapes filament morphology in angiosperms. Sci. Rep. 2014, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, T.; Ito, M.; Kato, M. YABBY2-homologue expression in lateral organs of Amborella trichopoda (Amborellaceae). Int. J. Plant Sci. 2004, 165, 917–924. [Google Scholar] [CrossRef]
- Morioka, K.; Yockteng, R.; Almeida, A.M.R.; Specht, C.D. Loss of YABBY2-like gene expression may underlie the evolution of the laminar style in Canna and contribute to floral morphological diversity in the Zingiberales. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Juarez, M.T.; Twigg, R.W.; Timmermans, M.C.P. Specification of adaxial cell fate maize leaf development. Development 2004, 131, 4533–4544. [Google Scholar] [CrossRef] [Green Version]
- Cai, J.; Liu, X.; Vanneste, K.; Proost, S.; Tsai, W.-C.; Liu, K.-W.; Chen, L.-J.; He, Y.; Xu, Q.; Bian, C.; et al. The genome sequence of the orchid Phalaenopsis equestris. Nat. Genet. 2014, 47, 65. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.Y.; Hsiao, Y.Y.; Chang, S.B.; Zhang, D.; Lan, S.R.; Liu, Z.J.; Tsai, W.C. Genome-wide identification of YABBY genes in Orchidaceae and their expression patterns in Phalaenopsis orchid. Genes 2020, 11, 955. [Google Scholar] [CrossRef]
- Toriba, T.; Harada, K.; Takamura, A.; Nakamura, H.; Ichikawa, H.; Suzaki, T.; Hirano, H.Y. Molecular characterization the YABBY gene family in Oryza sativa and expression analysis of OsYABBY1. Mol. Genet. Genom. 2007, 277, 457–468. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Nagasawa, N.; Kawasaki, S.; Matsuoka, M.; Nagato, Y.; Hirano, H.Y. The YABBY Gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 2004, 16, 500–509. [Google Scholar] [CrossRef] [Green Version]
- McAbee, J.M.; Hill, T.A.; Skinner, D.J.; Izhaki, A.; Hauser, B.A.; Meister, R.J.; Venugopala Reddy, G.; Meyerowitz, E.M.; Bowman, J.L.; Gasser, C.S. ABERRANT TESTA SHAPE encodes a KANADI family member, linking polarity determination to separation and growth of Arabidopsis ovule integuments. Plant J. 2006, 46, 522–531. [Google Scholar] [CrossRef] [Green Version]
- Villanueva, J.M.; Broadhvest, J.; Hauser, B.A.; Meister, R.J.; Schneitz, K.; Gasser, C.S. INNER NO OUTER regulates abaxial-adaxial patterning in Arabidopsis ovules. Genes Dev. 1999, 13, 3160–3169. [Google Scholar] [CrossRef] [Green Version]
- Filyushin, M.A.; Slugina, M.A.; Pyshnaya, O.N.; Kochieva, E.Z.; Shchennikova, A.V. Structure analysis of INNER NO OUTER (INO) homologs in Capsicum species. Russ. J. Genet. 2018, 54, 753–757. [Google Scholar] [CrossRef]
- Tang, H.; Lyons, E.; Schnable, J.C. Early history of the angiosperms. Adv. Bot. Res. 2014, 69, 195–222. [Google Scholar] [CrossRef]
- Chen, J.; Hao, Z.; Guang, X.; Zhao, C.; Wang, P.; Xue, L.; Zhu, Q.; Yang, L.; Sheng, Y.; Zhou, Y.; et al. Liriodendron genome sheds light on angiosperm phylogeny and species–pair differentiation. Nat. Plants 2019, 5, 18–25. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.P.; Sun, W.H.; Xiong, Y.F.; Jiang, Y.T.; Liu, X.D.; Liao, X.Y.; Zhang, D.Y.; Jiang, S.Z.; Li, Y.; Liu, B.; et al. The Phoebe genome sheds light on the evolution of magnoliids. Hortic. Res. 2020, 7, 146. [Google Scholar] [CrossRef]
- Lora, J.; Hormaza, J.I.; Herrero, M.; Gasser, C.S. Seedless fruits and the disruption of a conserved genetic pathway in angiosperm ovule development. Proc. Natl. Acad. Sci. USA 2011, 108, 5461–5465. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.N.; Jiang, Y.G.; Liu, D.; Ma, J.; Li, J.; Li, M.; Sui, S. Floral scent emission from nectaries in the adaxial side of the innermost and middle petals in Chimonanthus praecox. Int. J. Mol. Sci. 2018, 19, 3278. [Google Scholar] [CrossRef] [Green Version]
- Chaw, S.M.; Liu, Y.C.; Wu, Y.W.; Wang, H.Y.; Ling, C.Y.; Wu, C.S.; Ke, H.-M.; Chang, L.-Y.; Hsu, C.-Y.; Yang, H.-T.; et al. Stout camphor tree genome fills gaps in understanding of flowering plant genome evolution. Nat. Plants 2019, 5, 63. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Li, Z.; Zhao, Y.; Gao, M.; Wang, J.-Y.; Liu, K.W.; Wang, X.; Wu, L.W.; Jiao, Y.L.; Xu, Z.L.; et al. The Litsea genome and the evolution of the laurel family. Nat. Commun. 2020, 11, 1675. [Google Scholar] [CrossRef] [Green Version]
- Hu, L.; Xu, Z.; Wang, M.; Fan, R.; Yuan, D.; Wu, B.; Wu, H.; Qin, X.; Yan, L.; Tan, L.; et al. The chromosome-scale reference genome of black pepper provides insight into piperine biosynthesis. Nat. Commun. 2019, 10, 4702. [Google Scholar] [CrossRef]
- Lv, Q.; Qiu, J.; Liu, J.; Li, Z.; Zhang, W.; Wang, Q.; Fang, J.; Pan, J.; Chen, Z.; Cheng, W.; et al. The Chimonanthus salicifolius genome provides insight into magnoliid evolution and flavonoid biosynthesis. Plant J. 2020, 103, 1910–1923. [Google Scholar] [CrossRef]
- Shang, J.; Tian, J.; Cheng, H.; Yan, Q.; Chen, L. The chromosome-level wintersweet (Chimonanthus praecox) genome provides insights into floral scent biosynthesis and flowering in winter. Genome Biol. 2020, 21, 1. [Google Scholar] [CrossRef]
- Tang, Y.; Bao, X.; Liu, K.; Wang, J.; Zhang, J.; Feng, Y.; Wang, Y.; Lin, L.; Feng, J.; Li, C. Genome-wide identification and expression profiling of the auxin response factor (ARF) gene family in physic nut. PLoS ONE 2018, 13, e0201024. [Google Scholar] [CrossRef] [Green Version]
- Sawa, S.; Watanabe, K.; Goto, K.; Kanaya, E.; Morita, E.H.; Okada, K. FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 1999, 13, 1079–1088. [Google Scholar] [CrossRef] [Green Version]
- Bonaccorso, O.; Lee, J.E.; Puah, L.; Scutt, C.P.; Golz, J.F. FILAMENTOUS FLOWER controls lateral organ development by acting as both an activator and a repressor. BMC Plant Biol. 2012, 12, 176. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; Liu, X.; Yu, C.; Chen, Y.; Tang, H.; Zhang, L. Water lilies as emerging models for Darwin’s abominable mystery. Hortic. Res. 2017, 4. [Google Scholar] [CrossRef] [Green Version]
- Rudall, P.J.; Bateman, R.M. Roles of synorganisation, zygomorphy and heterotopy in floral evolution: The gynostemium and labellum of orchids and other lilioid monocots. Biol. Rev. Camb. Philos. Soc. 2002, 77, 403–441. [Google Scholar] [CrossRef]
- Tanaka, W.; Toriba, T.; Hirano, H. Three TOB1-related YABBY genes are required to maintain proper function of the spikelet and branch meristems in rice. New Phytol. 2017, 215, 2. [Google Scholar] [CrossRef] [Green Version]
- Crooks, G.; Hon, G.; Chandonia, J.; Brenner, S. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, 202–208. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]









| Category | C. micranthum | L. cubeba | P. bournei | L. chinense | C. praecox | C. salicifolius | P. nigrum |
|---|---|---|---|---|---|---|---|
| CRC/DL-like | 1 | 1 | 1 | 0 | 0 | 1 | 1 |
| INO-like | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| YAB2-like | 1 | 1 | 1 | 1 | 1 | 1 | 2 |
| FIL-like | 2 | 2 | 2 | 2 | 1 | 1 | 4 |
| YAB5-like | 3 | 3 | 3 | 2 | 2 | 2 | 7 |
| Total | 8 | 8 | 7 | 6 | 5 | 6 | 15 |
| Clade | Species | Gene Name | ORF (bp) | Protein Length (aa) | pI | Mv (Da) | Exon 1 | Exon 2 | Exon 3 | Exon 4 | Exon 5 | Exon 6 | Exon 7 | Exon 8 | Exon 9 | Exon 10 | Exon 11 | Exon 12 | Exon 13 | Exon 14 | Exon 15 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CRC/DL | Chimonanthus salicifolius | ChsCRC | 534 | 177 | 7.05 | 20,169.74 | 47 | 68 | 75 | 48 | 117 | 119 | 74 | ||||||||
| CRC/DL | Cinnamomum micranthum | CmCRC | 423 | 140 | 9.04 | 15,858.01 | 68 | 128 | 90 | 48 | 75 | 8 | |||||||||
| CRC/DL | Litsea cubeba | LcCRC | 468 | 155 | 8.98 | 17,773.17 | 14 | 14 | 128 | 128 | 90 | 90 | 48 | 48 | 75 | 75 | 107 | 107 | |||
| CRC/DL | Phoebe bournei | PboCRC | 1179 | 392 | 6.44 | 43,578.13 | 14 | 65 | 90 | 48 | 75 | 89 | 70 | 54 | 167 | 96 | 69 | 146 | 21 | 38 | 122 |
| CRC/DL | Piper nigrum | PnCRC | 519 | 172 | 9.40 | 19,208.75 | 71 | 131 | 90 | 48 | 75 | 62 | 35 | ||||||||
| CRC/DL | Chimonanthus praecox | ChpCRC | 519 | 172 | 8.82 | 19,406.98 | |||||||||||||||
| FIL | Litsea cubeba | LcFIL | 534 | 177 | 9.87 | 19,463.36 | 40 | 76 | 47 | 155 | 47 | 114 | 47 | 114 | 173 | 202 | 77 | 88 | |||
| FIL | Liriodendron chinense | LchiFIL | 615 | 204 | 8.37 | 22,893.1 | 68 | 74 | 75 | 48 | 75 | 114 | 101 | ||||||||
| FIL | Liriodendron chinense | LchiYAB3 | 639 | 212 | 7.64 | 23,455.73 | 68 | 74 | 120 | 48 | 75 | 155 | 92 | ||||||||
| FIL | Piper nigrum | PnYAB1 | 624 | 207 | 9.05 | 23,018.5 | 23 | 74 | 75 | 48 | 114 | 155 | 21 | 106 | |||||||
| FIL | Cinnamomum micranthum | CmYAB4 | 644 | 210 | 7.07 | 23,159.37 | 68 | 74 | 75 | 48 | 114 | 155 | 92 | ||||||||
| FIL | Phoebe bournei | PboFIL.1 | 636 | 211 | 7.64 | 23,189.42 | 68 | 77 | 75 | 48 | 114 | 155 | 92 | ||||||||
| FIL | Phoebe bournei | PboFIL.2 | 570 | 189 | 7.15 | 20,975.73 | 59 | 68 | 74 | 48 | 75 | 114 | 125 | ||||||||
| FIL | Cinnamomum micranthum | CmYAB1 | 636 | 211 | 7.1 | 23,241.44 | 68 | 77 | 75 | 48 | 114 | 155 | 92 | ||||||||
| FIL | Piper nigrum | PnFIL.1 | 636 | 211 | 6.78 | 23,373.68 | 68 | 74 | 75 | 48 | 114 | 155 | 95 | ||||||||
| FIL | Piper nigrum | PnFIL.3 | 669 | 222 | 8.18 | 24,635.92 | 68 | 77 | 75 | 48 | 126 | 158 | 110 | ||||||||
| FIL | Piper nigrum | PnFIL.2 | 669 | 222 | 8.19 | 24,655.98 | 68 | 77 | 75 | 48 | 126 | 158 | 110 | ||||||||
| FIL | Litsea cubeba | LcFIL.1 | 717 | 238 | 7.07 | 26455.15 | 68 | 77 | 114 | 48 | 75 | 155 | 173 | ||||||||
| FIL | Chimonanthus salicifolius | ChsFIL | 663 | 119 | 7.62 | 12,999.94 | 68 | 74 | 75 | 48 | 117 | 155 | 64 | 54 | |||||||
| FIL | Chimonanthus praecox | ChpFIL | 636 | 211 | 8.24 | 23,362.58 | |||||||||||||||
| INO | Liriodendron chinense | LchiINO | 264 | 87 | 5.33 | 9615.4 | 86 | 176 | |||||||||||||
| INO | Liriodendron chinense | LchiINO | 264 | 87 | 5.33 | 9615.4 | 86 | 176 | |||||||||||||
| INO | Cinnamomum micranthum | CmINO | 546 | 181 | 6.38 | 20,548.4 | 38 | 134 | 99 | 51 | 75 | 92 | 50 | ||||||||
| INO | Chimonanthus salicifolius | ChsYAB4 | 555 | 184 | 5.63 | 20,785.38 | 80 | 122 | 120 | 48 | 75 | 92 | 11 | ||||||||
| INO | Piper nigrum | PnINO | 615 | 204 | 4.88 | 22,845.96 | 98 | 134 | 120 | 48 | 75 | 35 | 98 | ||||||||
| YAB2 | Liriodendron chinense | LchiYAB2 | 495 | 164 | 8.8 | 18,231.7 | 23 | 116 | 114 | 48 | 75 | 113 | |||||||||
| YAB2 | Chimonanthus praecox | ChpYAB2 | 546 | 181 | 8.57 | 20,113.94 | |||||||||||||||
| YAB2 | Phoebe bournei | LcYAB2 | 540 | 179 | 7.61 | 20,284.98 | 68 | 119 | 111 | 48 | 75 | 113 | |||||||||
| YAB2 | Cinnamomum micranthum | CmYAB2 | 540 | 179 | 7.03 | 20,329.98 | 68 | 119 | 111 | 48 | 75 | 113 | |||||||||
| YAB2 | Chimonanthus salicifolius | ChsYAB2 | 555 | 184 | 9.10 | 20,908.74 | 68 | 119 | 117 | 48 | 75 | 122 | |||||||||
| YAB2 | Piper nigrum | PnYAB2 | 573 | 190 | 8.79 | 21,523.58 | 68 | 122 | 126 | 48 | 75 | 128 | |||||||||
| YAB2 | Piper nigrum | PnYAB2.1 | 573 | 190 | 9.02 | 21,564.56 | 68 | 122 | 126 | 48 | 75 | 128 | |||||||||
| YAB2 | Phoebe bournei | PboYAB2 | 804 | 267 | 8.92 | 30,743.77 | 113 | 75 | 48 | 111 | 119 | 128 | 203 | ||||||||
| YAB5 | Phoebe bournei | PboYAB5.3 | 216 | 71 | 6.68 | 7689.93 | 74 | 140 | |||||||||||||
| YAB5 | Phoebe bournei | PboYAB5.2 | 387 | 129 | 5.41 | 14,506.45 | 65 | 125 | 119 | 74 | |||||||||||
| YAB5 | Phoebe bournei | PboYAB5.1 | 477 | 158 | 8.45 | 17,567.2 | 74 | 150 | 48 | 75 | 65 | 59 | |||||||||
| YAB5 | Litsea cubeba | LcYAB5.2 | 483 | 160 | 8.53 | 18,198.95 | 74 | 134 | 91 | 75 | 65 | 38 | |||||||||
| YAB5 | Litsea cubeba | LcYAB5 | 507 | 168 | 8.2 | 18,764.34 | 74 | 119 | 117 | 48 | 75 | 68 | |||||||||
| YAB5 | Litsea cubeba | LcYAB5.1 | 813 | 270 | 9.46 | 30,701.78 | 25 | 138 | 119 | 117 | 48 | 75 | 65 | 37 | 180 | ||||||
| YAB5 | Cinnamomum micranthum | CmYAB5.2 | 543 | 180 | 6.99 | 20,000.71 | 74 | 119 | 117 | 48 | 75 | 65 | 38 | ||||||||
| YAB5 | Cinnamomum micranthum | CmYAB5.1 | 771 | 183 | 9.26 | 20,592.77 | 74 | 149 | 117 | 48 | 75 | 65 | 17 | ||||||||
| YAB5 | Cinnamomum micranthum | CmYAB5 | 507 | 168 | 8.52 | 18,780.31 | 74 | 119 | 117 | 48 | 75 | 68 | |||||||||
| YAB5 | Liriodendron chinense | LchiYAB5.1 | 546 | 181 | 8.57 | 20,470.27 | 74 | 119 | 114 | 48 | 75 | 71 | 38 | ||||||||
| YAB5 | Liriodendron chinense | LchiYAB5 | 558 | 185 | 8.44 | 20,887.91 | 74 | 119 | 120 | 48 | 75 | 68 | 47 | ||||||||
| YAB5 | Chimonanthus praecox | ChpYAB5.1 | 555 | 184 | 6.99 | 20,693.56 | |||||||||||||||
| YAB5 | Chimonanthus praecox | ChpYAB5.2 | 552 | 183 | 8.6 | 20,694.87 | |||||||||||||||
| YAB5 | Chimonanthus salicifolius | ChsYAB5.1 | 555 | 184 | 6.99 | 20,724.53 | 74 | 119 | 117 | 48 | 75 | 68 | 47 | ||||||||
| YAB5 | Chimonanthus salicifolius | ChsYAB5.2 | 657 | 218 | 8.99 | 24,354.12 | 72 | 48 | 114 | 119 | 83 | 74 | 104 | 35 | |||||||
| YAB5 | Piper nigrum | PnYAB5.1 | 513 | 170 | 8.80 | 19,384.9 | 74 | 122 | 126 | 48 | 75 | 62 | |||||||||
| YAB5 | Piper nigrum | PnYAB5.2 | 513 | 170 | 8.80 | 19,384.9 | 74 | 122 | 126 | 48 | 75 | 62 | |||||||||
| YAB5 | Piper nigrum | PnYAB5.3 | 507 | 168 | 8.85 | 19,035.58 | 74 | 122 | 120 | 48 | 75 | 62 | |||||||||
| YAB5 | Piper nigrum | PnYAB5.4 | 681 | 226 | 8.55 | 24,924.55 | 74 | 122 | 135 | 48 | 75 | 72 | 148 | ||||||||
| YAB5 | Piper nigrum | PnYAB5.5 | 576 | 191 | 8.37 | 21,440.51 | 74 | 68 | 75 | 48 | 135 | 122 | 47 | ||||||||
| YAB5 | Piper nigrum | PnYAB5.6 | 681 | 226 | 8.71 | 24,969.68 | 74 | 122 | 135 | 48 | 75 | 72 | 148 | ||||||||
| YAB5 | Piper nigrum | PnYAB5.7 | 510 | 169 | 8.89 | 19,289.98 | 68 | 119 | 117 | 48 | 75 | 122 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Liao, X.-Y.; Zheng, Y.; Zhu, M.-J.; Yu, X.; Jiang, Y.-T.; Zhang, D.-Y.; Ma, L.; Xu, X.-Y.; Liu, Z.-J.; et al. Genome-Wide Identification of the YABBY Gene Family in Seven Species of Magnoliids and Expression Analysis in Litsea. Plants 2021, 10, 21. https://doi.org/10.3390/plants10010021
Liu X, Liao X-Y, Zheng Y, Zhu M-J, Yu X, Jiang Y-T, Zhang D-Y, Ma L, Xu X-Y, Liu Z-J, et al. Genome-Wide Identification of the YABBY Gene Family in Seven Species of Magnoliids and Expression Analysis in Litsea. Plants. 2021; 10(1):21. https://doi.org/10.3390/plants10010021
Chicago/Turabian StyleLiu, Xuedie, Xing-Yu Liao, Yu Zheng, Meng-Jia Zhu, Xia Yu, Yu-Ting Jiang, Di-Yang Zhang, Liang Ma, Xin-Yu Xu, Zhong-Jian Liu, and et al. 2021. "Genome-Wide Identification of the YABBY Gene Family in Seven Species of Magnoliids and Expression Analysis in Litsea" Plants 10, no. 1: 21. https://doi.org/10.3390/plants10010021