Targeted Liposomes Encapsulating miR-603 Complexes Enhance Radiation Sensitivity of Patient-Derived Glioblastoma Stem-Like Cells
Abstract
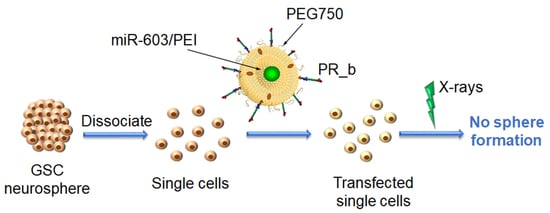
:1. Introduction
2. Materials and Methods
2.1. Materials and Cells
2.2. Preparation and Characterization of miR-603/PEI Complexes
2.3. Preparation and Characterization of Liposomes
2.4. Cryogenic Transmission Electron Microscopy (Cryo-TEM)
2.5. Expression of Integrin α5β1
2.6. Flow Cytometry
2.7. Confocal Microscopy
2.8. RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
- Hs_miR-603_3 miScript Primer (MS00037933, Qiagen, Germantown, MD, USA),
- IGF1: 5′-GCAGCACTCATCCACGATGC-3′ (forward primer),
- 5′-TGTGGAGACAGGGGCTTTTATTTC-3′ (reverse primer).
- IGF1R: 5′-AAGTTCTGGTTGTCGAGGA-3′ (forward primer),
- 5′-GAGCAGCTAGAAGGGAATTAC-3′ (reverse primer).
- 18s: 5′-TTGCCCTCCAATGGATCCT-3′ (forward primer),
- 5′-GGGAGGTAGTGACGAAAAATAACAAT-3′ (reverse primer).
2.9. Limiting Dilution Assay
3. Results and Discussion
3.1. Encapsulation of miR-603/PEI Complexes into PR_b Liposomes
3.2. Targeting Patient-Derived Glioblastoma Cell Lines with PR_b Liposomes
3.3. PR_b Liposomes Encapsulating miR-603/PEI Complexes Suppress the Expression of IGF1 and IGF1R in Patient-Derived Glioblastoma Cells
3.4. PR_b Liposomes Encapsulating miR-603/PEI Complexes Enhance Radiation Sensitivity of Patient-Derived Glioblastoma Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bartek, J.; Ng, K.; Bartek, J.; Fischer, W.; Carter, B.; Chen, C.C. Key concepts in glioblastoma therapy. J. Neurol. Neurosurg. Psychiatry 2012, 83, 753–760. [Google Scholar] [CrossRef]
- Lai, E.C. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 2002, 30, 363–364. [Google Scholar] [CrossRef]
- Garofalo, M.; Croce, C.M. microRNAs: Master Regulators as Potential Therapeutics in Cancer. Annu. Rev. Pharmacol. Toxicol. 2011, 51, 25–43. [Google Scholar] [CrossRef]
- Ramakrishnan, V.; Xu, B.; Akers, J.; Nguyen, T.; Ma, J.; Dhawan, S.; Ning, J.; Mao, Y.; Hua, W.; Kokkoli, E.; et al. Radiation-induced extracellular vesicle (EV) release of miR-603 promotes IGF1-mediated stem cell state in glioblastomas. EBioMedicine 2020, 55, 102736. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-K. RNA Therapy: Current Status and Future Potential. Chonnam Med. J. 2020, 56, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Gao, D.Y.; Huang, L. In vivo delivery of miRNAs for cancer therapy: Challenges and strategies. Adv. Drug Deliv. Rev. 2015, 81, 128–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scomparin, A.; Polyak, D.; Krivitsky, A.; Satchi-Fainaro, R. Achieving successful delivery of oligonucleotides—From physico-chemical characterization to in vivo evaluation. Biotechnol. Adv. 2015, 33, 1294–1309. [Google Scholar] [CrossRef]
- Levine, R.M.; Dinh, C.V.; Harris, M.A.; Kokkoli, E. Targeting HPV-infected cervical cancer cells with PEGylated liposomes encapsulating siRNA and the role of siRNA complexation with polyethylenimine. Bioeng. Transl. Med. 2016, 1, 168–180. [Google Scholar] [CrossRef]
- Campani, V.; Zappavigna, S.; Scotti, L.; Abate, M.; Porru, M.; Leonetti, C.; Caraglia, M.; De Rosa, G. Hybrid lipid self-assembling nanoparticles for brain delivery of microRNA. Int. J. Pharm. 2020, 588, 119693. [Google Scholar] [CrossRef]
- Bayraktar, R.; Pichler, M.; Kanlikilicer, P.; Ivan, C.; Bayraktar, E.; Kahraman, N.; Aslan, B.; Oguztuzun, S.; Ulasli, M.; Arslan, A.; et al. MicroRNA 603 acts as a tumor suppressor and inhibits triplenegative breast cancer tumorigenesis by targeting elongation factor 2 kinase. Oncotarget 2017, 8, 11641–11658. [Google Scholar] [CrossRef] [Green Version]
- Mardilovich, A.; Craig, J.A.; McCammon, M.Q.; Garg, A.; Kokkoli, E. Design of a novel fibronectin-mimetic peptide-amphiphile for functionalized biomaterials. Langmuir 2006, 22, 3259–3264. [Google Scholar] [CrossRef]
- Dontenwill, M.; Martin, S.; Janouskova, H. Integrins and p53 pathways in glioblastoma resistance to temozolomide. Front. Oncol. 2012, 2, 157. [Google Scholar]
- Vangala, V.; Nimmu, N.V.; Khalid, S.; Kuncha, M.; Sistla, R.; Banerjee, R.; Chaudhuri, A. Combating Glioblastoma by Codelivering the Small-Molecule Inhibitor of STAT3 and STAT3siRNA with α5β1 Integrin Receptor-Selective Liposomes. Mol. Pharm. 2020, 17, 1859–1874. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Tang, Z.; Li, Y.; Yin, G.; Liu, Z.; Gao, J.; Chen, Y.; Chen, F. Patient-derived xenografts of different grade gliomas retain the heterogeneous histological and genetic features of human gliomas. Cancer Cell Int. 2020, 20, 1. [Google Scholar] [CrossRef]
- Singh, S.K.; Hawkins, C.; Clarke, I.D.; Squire, J.A.; Bayani, J.; Hide, T.; Henkelman, R.M.; Cusimano, M.D.; Dirks, P.B. Identification of human brain tumour initiating cells. Nature 2004, 432, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Kozono, D.; Li, J.; Nitta, M.; Sampetrean, O.; Gonda, D.; Kushwaha, D.S.; Merzon, D.; Ramakrishnan, V.; Zhu, S.; Zhu, K.; et al. Dynamic epigenetic regulation of glioblastoma tumorigenicity through LSD1 modulation of MYC expression. Proc. Natl. Acad. Sci. USA 2015, 112, E4055–E4064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adil, M.M.; Erdman, Z.S.; Kokkoli, E. Transfection mechanisms of polyplexes, lipoplexes, and stealth liposomes in α5β1 integrin bearing DLD-1 colorectal cancer cells. Langmuir 2014, 30, 3802–3810. [Google Scholar] [CrossRef] [PubMed]
- Levine, R.M.; Kokkoli, E. Dual-ligand α5β1 and α6β4 integrin targeting enhances gene delivery and selectivity to cancer cells. J. Control. Release 2017, 251, 24–36. [Google Scholar] [CrossRef]
- Hu, Y.; Smyth, G.K. ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 2009, 347, 70–78. [Google Scholar] [CrossRef]
- Pangburn, T.O.; Bates, F.S.; Kokkoli, E. Polymersomes functionalized via “click” chemistry with the fibronectin mimetic peptides PR_b and GRGDSP for targeted delivery to cells with different levels of a5b1 expression. Soft Matter 2012, 8, 4449–4461. [Google Scholar] [CrossRef]
- Gong, J.; Wang, D.; Sun, L.; Zborowska, E.; Willson, J.K.; Brattain, M.G. Role of α5β1 integrin in determining malignant properties of colon carcinoma cells. Cell Growth Differ. 1997, 8, 83–90. [Google Scholar]
- Jayne, D.G.; Heath, R.M.; Dewhurst, O.; Scott, N.; Guillou, P.J. Extracellular matrix proteins and chemoradiotherapy: α5β1 integrin as a predictive marker in rectal cancer. Eur. J. Surg. Oncol. 2002, 28, 30–36. [Google Scholar] [CrossRef]
- Jia, Y.; Zeng, Z.-Z.; Markwart, S.M.; Rockwood, K.F.; Ignatoski, K.M.W.; Ethier, S.P.; Livant, D.L. Integrin fibronectin receptors in matrix metalloproteinase-1–dependent invasion by breast cancer and mammary epithelial cells. Cancer Res. 2004, 64, 8674–8681. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Bell, K.; Mousa, S.A.; Varner, J.A. Regulation of angiogenesis in vivo by ligation of integrin α5β11 with the central cell-binding domain of fibronectin. Am. J. Pathol. 2000, 156, 1345–1362. [Google Scholar] [CrossRef]
- Maglott, A.; Bartik, P.; Cosgun, S.; Klotz, P.; Rondé, P.; Fuhrmann, G.; Takeda, K.; Martin, S.; Dontenwill, M. The Small α5β1 Integrin Antagonist, SJ749, Reduces Proliferation and Clonogenicity of Human Astrocytoma Cells. Cancer Res. 2006, 66, 6002–6007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dingemans, A.-M.C.; van den Boogaart, V.; Vosse, B.A.; van Suylen, R.-J.; Griffioen, A.W.; Thijssen, V.L. Integrin expression profiling identifies integrin alpha5 and beta1 as prognostic factors in early stage non-small cell lung cancer. Mol. Cancer 2010, 9, 152. [Google Scholar] [CrossRef] [Green Version]
- Muschler, J.L.; Horwitz, A.F. Down-regulation of the chicken α5β1 integrin fibronectin receptor during development. Development 1991, 113, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Shi, F.; Sottile, J. Caveolin-1-dependent beta1 integrin endocytosis is a critical regulator of fibronectin turnover. J. Cell Sci. 2008, 121, 2360–2371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, R.D.; Marks, D.L.; Holicky, E.L.; Wheatley, C.L.; Kaptzan, T.; Sato, S.B.; Kobayashi, T.; Ling, K.; Pagano, R.E. Gangliosides and β1-Integrin Are Required for Caveolae and Membrane Domains. Traffic 2010, 11, 348–360. [Google Scholar] [CrossRef] [Green Version]
- Boussif, O.; Lezoualc’h, F.; Zanta, M.a.; Mergny, M.D.; Scherman, D.; Demeneix, B.; Behr, J.P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine. Proc. Natl. Acad. Sci. USA 1995, 92, 7297–7301. [Google Scholar] [CrossRef] [Green Version]
- Akinc, A.; Thomas, M.; Klibanov, A.M.; Langer, R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J. Gene Med. 2005, 7, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Benjaminsen, R.V.; Mattebjerg, M.A.; Henriksen, J.R.; Moghimi, S.M.; Andresen, T.L. The Possible “Proton Sponge” Effect of Polyethylenimine (PEI) Does Not Include Change in Lysosomal pH. Mol. Ther. 2013, 21, 149–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Formulation | Encapsulating Load | Diameter (nm) 1 | PDI 2 | Zeta Potential (mV) | Encapsulation Efficiency (%) |
|---|---|---|---|---|---|
| Non-Targeted Liposomes | miR-603 | 174.5 ± 2.0 | 0.025 ± 0.006 | −25.8 ± 0.5 | 70 ± 4 |
| Non-Targeted Liposomes | miR-603/PEI | 173.6 ± 0.7 | 0.068 ± 0.008 | −24.7 ± 2.2 | 44 ± 5 |
| PR_b Liposomes | miR-603 | 176.8 ± 1.5 | 0.029 ± 0.002 | 6.3 ± 0.1 | 69 ± 7 |
| PR_b Liposomes | miR-603/PEI | 173.2 ± 3.8 | 0.036 ± 0.030 | 4.0 ± 1.3 | 58 ± 3 |
| Treatment | 24 h Liposomal Incubation | 95% CI 1 for 24 h Incubation | 48 h Liposomal Incubation | 95% CI 1 for 48 h Incubation |
|---|---|---|---|---|
| PBS Control | 0.38 | (0.30, 0.47) | 0.37 | (0.28, 0.48) |
| PBS Control + IR | 0.09 | (0.07, 0.10) | 0.09 | (0.07, 0.11) |
| PR_b Liposomes | 0.35 | (0.28, 0.44) | 0.36 | (0.27, 0.47) |
| PR_b Liposomes + IR | 0.08 | (0.07, 0.10) | 0.09 | (0.07, 0.12) |
| PR_b Liposomes w/miR-603 | 0.23 | (0.19, 0.29) | 0.25 | (0.19, 0.32) |
| PR_b Liposomes w/miR-603 + IR | 0.07 | (0.06, 0.09) | 0.07 | (0.05, 0.09) |
| PR_b liposomes w/miR-603/PEI | 0.08 | (0.06, 0.10) | 0.08 | (0.06, 0.11) |
| PR_b liposomes w/miR-603/PEI + IR | 0.03 | (0.03, 0.04) | 0.03 | (0.03, 0.05) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shabana, A.M.; Xu, B.; Schneiderman, Z.; Ma, J.; Chen, C.C.; Kokkoli, E. Targeted Liposomes Encapsulating miR-603 Complexes Enhance Radiation Sensitivity of Patient-Derived Glioblastoma Stem-Like Cells. Pharmaceutics 2021, 13, 1115. https://doi.org/10.3390/pharmaceutics13081115
Shabana AM, Xu B, Schneiderman Z, Ma J, Chen CC, Kokkoli E. Targeted Liposomes Encapsulating miR-603 Complexes Enhance Radiation Sensitivity of Patient-Derived Glioblastoma Stem-Like Cells. Pharmaceutics. 2021; 13(8):1115. https://doi.org/10.3390/pharmaceutics13081115
Chicago/Turabian StyleShabana, Ahmed M., Beibei Xu, Zachary Schneiderman, Jun Ma, Clark C. Chen, and Efrosini Kokkoli. 2021. "Targeted Liposomes Encapsulating miR-603 Complexes Enhance Radiation Sensitivity of Patient-Derived Glioblastoma Stem-Like Cells" Pharmaceutics 13, no. 8: 1115. https://doi.org/10.3390/pharmaceutics13081115