Determination of the Infectious Agent of Translucent Post-Larva Disease (TPD) in Penaeus vannamei
Abstract
:1. Introduction
2. Results
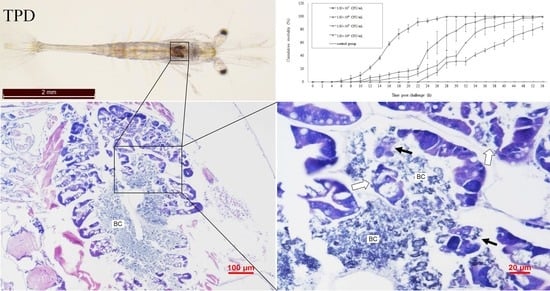
2.1. Clinical Signs
2.2. Detection of Known Pathogens in the Diseased Samples
2.3. Bacterial Identification
2.4. Virulence Genes in the Isolated Strain
2.5. Pathogenicity of the Vibrio. Parahaemolyticus Determined by the Challenge Test
2.6. Histopathology Analysis of the Vibrio Infection
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Experiment Shrimp
4.2. Detection of Eight Known Shrimp Pathogens in the Diseased Samples
4.3. Bacteria Isolation
4.4. Bacteria Identification
4.5. Analysis of Virulent Genes in the Isolated Strain
4.6. Experimental Challenge by Immersion
4.7. Histopathology
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kibenge, F.S.B. Emerging viruses in aquaculture. Curr. Opin. Virol. 2019, 34, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Tandel, G.M.; John, K.R.; George, M.R.; Jeyaseelan, M.J.P. Current status of viral diseases in Indian shrimp aquaculture. Acta Virol. 2017, 61, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Liu, Q.; Liu, S.; Yang, H.; Liu, S.; Zhu, L.; Yang, B.; Jin, J.; Ding, L.; Wang, X.; et al. A new nodavirus is associated with covert mortality disease of shrimp. J. Gen. Virol. 2014, 95, 2700–2709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Xu, T.; Wan, X.; Liu, S.; Wang, X.; Li, X.; Dong, X.; Yang, B.; Huang, J. Prevalence and distribution of covert mortality nodavirus (CMNV) in cultured crustacean. Virus Res. 2017, 233, 113–119. [Google Scholar] [CrossRef]
- Qiu, L.; Chen, M.-M.; Wan, X.Y.; Zhang, Q.; Li, C.; Dong, X.; Yang, B.; Huang, J. Detection and quantification of shrimp hemocyte iridescent virus by TaqMan probe based real-time PCR. J. Invertebr. Pathol. 2018, 154, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Harkell, L. Shrimp Hatcheries in China Hit by ‘Glass Post-Larvae’. Undercurrentnews, 2020-4-22. 2020. Available online: https://www.undercurrentnews.com/2020/04/22/shrimp-hatcheries-in-china-hit-by-glass-post-larvae/ (accessed on 23 April 2020).
- Harkell, L. Chinese Scientists Confirm New Virus Causes Shrimp ‘Glass Post-Larvae’. 2020-5-8. 2020. Available online: https://www.undercurrentnews.com/2020/05/08/Chinese-scientists-confirm-new-virus-causes-shrimp-glass-post-larvae/ (accessed on 8 May 2020).
- Pascual, J.; Macián, M.C.; Arahal, D.R.; Garay, E.; Pujalte, M.J. Multilocus sequence analysis of the central clade of the genus Vibrio by using the 16S rRNA, recA, pyrH, rpoD, gyrB, rctB and toxR genes. Int. J. Syst. Evol. Microbiol. 2010, 60, 154–165. [Google Scholar] [CrossRef] [Green Version]
- Flegel, T.W. Historic emergence, impact and current status of shrimp pathogens in Asia. J. Invertebr. Pathol. 2012, 110, 166–173. [Google Scholar] [CrossRef]
- Yong, C.Y.; Yeap, S.K.; Omar, A.R.; Tan, W.S. Advances in the study of nodavirus. PeerJ 2017, 5, 3841. [Google Scholar] [CrossRef]
- Zhang, B.C.; Liu, F.; Bian, H.H.; Liu, J.; Pan, L.Q.; Huang, J. Isolation, identification, and pathogenicity analysis of a Vibrio parahaemolyticus strain from Litopenaeus vannamei. Prog Fish Sci. 2012, 33, 56–62. (In Chinese) [Google Scholar] [CrossRef]
- Kondo, H.; Tinwongger, S.; Proespraiwong, P.; Mavichak, R.; Unajak, S.; Nozaki, R.; Hirono, I. Draft Genome Sequences of Six Strains of Vibrio parahaemolyticus Isolated from Early Mortality Syndrome/Acute Hepatopancreatic Necrosis Disease Shrimp in Thailand. Genome Announc. 2014, 2, e00221-14. [Google Scholar] [CrossRef] [Green Version]
- Tran, L.; Nunan, L.; Redman, R.; Mohney, L.; Pantoja, C.R.; Fitzsimmons, K.; Lightner, D. Determination of the infectious nature of the agent of acute hepatopancreatic necrosis syndrome affecting penaeid shrimp. Dis. Aquat. Org. 2013, 105, 45–55. [Google Scholar] [CrossRef]
- De La Pena, L.; Cabillon, N.; Catedral, D.; Amar, E.; Usero, R.; Monotilla, W.; Calpe, A.; Fernandez, D.; Saloma, C. Acute hepatopancreatic necrosis disease (AHPND) outbreaks in Penaeus vannamei and P. monodon cultured in the Philippines. Dis. Aquat. Org. 2015, 116, 251–254. [Google Scholar] [CrossRef] [PubMed]
- Chonsin, K.; Matsuda, S.; Theethakaew, C.; Junjhon, J.; Suthienkul, O.; Kodama, T.; Suzuki, Y.; Iida, T. Genetic diversity of Vibrio parahaemolyticus strains isolated from farmed Pacific white shrimp and ambient pond water affected by acute hepatopancreatic necrosis disease outbreak in Thailand. FEMS Microbiol. Lett. 2015, 363, 222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhar, A.K.; Piamsomboon, P.; Caro, L.F.A.; Kanrar, S.; Adami, R.; Juan, Y.-S. First report of acute hepatopancreatic necrosis disease (AHPND) occurring in the USA. Dis. Aquat. Org. 2019, 132, 241–247. [Google Scholar] [CrossRef]
- Restrepo, L.; Bayot, B.; Arciniegas, S.; Bajaña, L.; Betancourt, I.; Panchana, F.; Muñoz, A.R. PirVP genes causing AHPND identified in a new Vibrio species (Vibrio punensis) within the commensal Orientalis clade. Sci. Rep. 2018, 8, 13080. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.; Bondad-Reantaso, M. Impacts of acute hepatopancreatic necrosis disease on commercial shrimp aquaculture. Rev. Sci. Tech. 2019, 38, 477–490. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Wei, D.; Yang, Q.; Xie, G.; Pang, B.; Wang, Y.; Lan, R.; Wang, Q.; Dong, X.; Zhang, X.; et al. Horizontal Plasmid Transfer Promotes the Dissemination of Asian Acute Hepatopancreatic Necrosis Disease and Provides a Novel Mechanism for Genetic Exchange and Environmental Adaptation. mSystems 2020, 5, 00799-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.T.; Chen, I.T.; Lee, C.T.; Chen, C.Y.; Lin, S.S.; Hor, L.I.; Tseng, T.C.; Huang, Y.T.; Sritunyalucksana, K.; Thitamadee, S.; et al. Draft Genome Sequences of Four Strains of Vibrio parahaemolyticus, Three of Which Cause Early Mortality Syndrome/Acute Hepatopancreatic Necrosis Disease in Shrimp in China and Thailand. Genome Announc. 2014, 2, e00816-14. [Google Scholar] [CrossRef] [Green Version]
- Han, J.E.; Tang, K.F.J.; Tran, L.H.; Lightner, D.V. Photorhabdus insect-related (Pir) toxin-like genes in a plasmid of Vibrio parahaemolyticus, the causative agent of acute hepatopancreatic necrosis disease (AHPND) of shrimp. Dis. Aquat. Org. 2015, 113, 33–40. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.T.; Chen, I.T.; Yang, Y.T.; Ko, T.P.; Huang, Y.T.; Huang, J.Y.; Huang, M.-F.; Lin, S.J.; Chen, C.Y.; Lin, S.-S.; et al. The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin. Proc. Natl. Acad. Sci. USA 2015, 112, 10798–10803. [Google Scholar] [CrossRef] [Green Version]
- Kongrueng, J.; Yingkajorn, M.; Bunpa, S.; Sermwittayawong, N.; Singkhamanan, K.; Vuddhakul, V. Characterization of Vibrio parahaemolyticus causing acute hepatopancreatic necrosis disease in southern Thailand. J. Fish Dis. 2014, 38, 957–966. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Tang, K.; Lightner, D. Genotyping of virulence plasmid from Vibrio parahaemolyticus isolates causing acute hepatopancreatic necrosis disease in shrimp. Dis. Aquat. Org. 2015, 115, 245–251. [Google Scholar] [CrossRef]
- Kumar, V.; De Bels, L.; Couck, L.; Baruah, K.; Bossier, P.; Broeck, W.V.D. PirABVP Toxin Binds to Epithelial Cells of the Digestive Tract and Produce Pathognomonic AHPND Lesions in Germ-Free Brine Shrimp. Toxins 2019, 11, 717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, R.; Ng, T.H.; Chang, C.; Tung, T.; Lin, S.; Lo, C.; Wang, H.C. Bile acid and bile acid transporters are involved in the pathogenesis of acute hepatopancreatic necrosis disease in white shrimp Litopenaeus vannamei. Cell. Microbiol. 2020, 22, e13127. [Google Scholar] [CrossRef] [PubMed]
- Phiwsaiya, K.; Charoensapsri, W.; Taengphu, S.; Dong, H.T.; Sangsuriya, P.; Nguyen, G.T.T.; Pham, H.Q.; Amparyup, P.; Sritunyalucksana, K.; Taengchaiyaphum, S.; et al. A Natural Vibrio parahaemolyticus ΔpirAVp pirBVp+ mutant kills shrimp but produces neither PirVp toxins nor acute hepatopancreatic necrosis disease lesions. Appl Environ Microbiol. 2017, 83, e00680-17. [Google Scholar] [CrossRef] [Green Version]
- Han, J.E.; Tang, F.J.; Aranguren, L.F.; Piamsomboonb, P. Characterization and pathogenicity of acute hepatopancreatic necrosis disease natural mutants, pirABvp (-) V. parahaemolyticus, and pirABvp (+) V. campbellii strains. Aquaculture 2017, 470, 84–90. [Google Scholar] [CrossRef]
- Andrea, V.; Suwimon, T.; Armando, L.H.; Chìo, M.M.; Ha Thanh, D.; Saengchan, S. Detection of Vibrio campbellii and V. parahaemolyticus carrying full-length pirABVp but only V. campbellii produces PirVp toxins. Aquaculture 2020, 519, 734708. [Google Scholar] [CrossRef]
- Victorio-De, L.M.; Vibanco-Pérez, N.; Soto-Rodriguez, S.; Pereyra, A.; Zenteno, E.; Cano-Sánchez, P. The B subunit of PirABvp toxin secreted from Vibrio parahaemolyticus causing AHPND is an amino sugar specific lectin. Pathogens 2020, 9, 182. [Google Scholar] [CrossRef] [Green Version]
- Robertson, P.; Calderón, J.; Carrera, L.; Stark, J.; Zherdmant, M.; Austin, B. Experimental Vibrio harveyi infections in Penaeus vannamei larvae. Dis. Aquat. Org. 1998, 32, 151–155. [Google Scholar] [CrossRef] [Green Version]
- Vandenberghe, J.; Verdonck, L.; Robles, R.; Rivera, G.; Bolland, A.; Balladares, M.; Gómez-Gil, B.; Calderon, J.; Sorgeloos, P.; Swings, J. Vibrios Associated with Litopenaeus vannamei Larvae, Postlarvae, Broodstock, and Hatchery Probionts. Appl. Environ. Microbiol. 1999, 65, 2592–2597. [Google Scholar] [CrossRef] [Green Version]
- World Organization for Animal Health (OIE). Manual of Diagnostic Tests for Aquatic Animals; OIE: Paris, France, 2019; Chapter 2.3.1.2. [Google Scholar]
- Jaroenlak, P.; Sanguanrut, P.; Williams, T.A.; Stentiford, G.D.; Flegel, T.W.; Sritunyalucksana, K.; Itsathitphaisarn, O. A Nested PCR Assay to Avoid False Positive Detection of the Microsporidian Enterocytozoon hepatopenaei (EHP) in Environmental Samples in Shrimp Farms. PLoS ONE 2016, 11, e0166320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, C.S.; Handley, K.M.; Wrighton, K.; Frischkorn, K.R.; Thomas, B.C.; Banfield, J.F. Short-Read Assembly of Full-Length 16S Amplicons Reveals Bacterial Diversity in Subsurface Sediments. PLoS ONE 2013, 8, e56018. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.L.; Lee, C.C.; Lin, Y.-L.; Yin, K.-M.; Ho, C.-L.; Liu, T. Obtaining long 16S rDNA sequences using multiple primers and its application on dioxin-containing samples. BMC Bioinform. 2015, 16, S13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dangtip, S.; Sirikharin, R.; Sanguanrut, P.; Thitamadee, S.; Sritunyalucksana, K.; Taengchaiyaphum, S.; Mavichak, R.; Proespraiwong, P.; Flegel, T.W. AP4 method for two-tube nested PCR detection of AHPND isolates of Vibrio parahaemolyticus. Aquac. Rep. 2015, 2, 158–162. [Google Scholar] [CrossRef] [Green Version]
- Garrity, G.M.; Bell, J.A.; Lilburn, T.G. Bergey’s Manual of Systematic Bacteriology; Science Press: New York, NY, USA, 2004. [Google Scholar]
- Teng, T.; Liang, L.; Chen, K.; Xi, B.W.; Xie, J.; Xu, P. Isolation, identification and phenotypic and molecular characterization of pathogenic Vibrio vulnificus isolated from Litopenaeus vannamei. PLoS ONE 2017, 12, e0186135. [Google Scholar] [CrossRef]
- Lightner, D.V. A Handbook of Shrimp Pathology and Diagnostic Procedures for Diseases of Cultured Penaeid Shrimp; World Aquaculture Society: Baton Rouge, LA, USA, 1996. [Google Scholar]








| Gene | Primer | Primer Sequences (5′-3′) | References |
|---|---|---|---|
| 16S rDNA | 27F | AGAGTTTGATCMTGGCTCAG | [35,36] |
| 1492R | GGYTACCTTGTTACGACTT | ||
| rpoD | rpoD-F | ACGACTGACCCGGTACGCATGTAYATGMGNGARATGGGNACNGT | [8] |
| rpoD-R | ATAGAAATAACCAGACGTAAGTTNGCYTCNACCATYTCYTTYT | ||
| rctB | rctB -F | ATHGARTTYACNGAYTTYCARYTNCAY | |
| rctB-R | YTTNCTYTGHATNGGYTCRAAYTCNCCRTC | ||
| toxR | toxR-F | GANCARGGNTTYGARGTNGAYGAYTC | |
| toxR-R | TTDKKTTGNCCNCYNGTVGCDATNAC | ||
| ldh | ldh-F | AAAGCGGATTATGCAGAAGCACTG | [29] |
| ldh-R | GCTACTTTCTAGCATTTTCTCTGC | ||
| pirAVp | pirA-284F | TGACTATTCTCACGATTGGACTG | [21] |
| pirA-284R | CACGACTAGCGCCATTGTTA | ||
| pirBVp | pirB-392F | TGATGAAGTGATGGGTGCTC | |
| pirB-392R | TGTAAGCGCCGTTTAACTCA | ||
| pirABVp | pirAB2020-F | GCACCGTAAATTTTCAGGTT | [27] |
| pirAB2020-R | CGTTGCAATCTAAGACATAG | ||
| AP4 | AP4-1 | AP4-F1: ATGAGTAACAATATAAAACATGAAAC | [37] |
| AP4-R1: ACGATTTCGACGTTCCCCAA | |||
| AP4-2 | AP4-F2: TTGAGAATACGGGACGTGGG | ||
| AP4-R2: GTTAGTCATGTGAGCACCTTC |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, Y.; Xie, G.; Jia, T.; Xu, T.; Wang, C.; Wan, X.; Li, Y.; Luo, K.; Bian, X.; Wang, X.; et al. Determination of the Infectious Agent of Translucent Post-Larva Disease (TPD) in Penaeus vannamei. Pathogens 2020, 9, 741. https://doi.org/10.3390/pathogens9090741
Zou Y, Xie G, Jia T, Xu T, Wang C, Wan X, Li Y, Luo K, Bian X, Wang X, et al. Determination of the Infectious Agent of Translucent Post-Larva Disease (TPD) in Penaeus vannamei. Pathogens. 2020; 9(9):741. https://doi.org/10.3390/pathogens9090741
Chicago/Turabian StyleZou, Ying, Guosi Xie, Tianchang Jia, Tingting Xu, Chong Wang, Xiaoyuan Wan, Yingxia Li, Kun Luo, Xiaodong Bian, Xiuhua Wang, and et al. 2020. "Determination of the Infectious Agent of Translucent Post-Larva Disease (TPD) in Penaeus vannamei" Pathogens 9, no. 9: 741. https://doi.org/10.3390/pathogens9090741