Biocidal Resistance in Clinically Relevant Microbial Species: A Major Public Health Risk
Abstract
:1. Introduction
2. Antimicrobial Biocide Use
3. Biocidal Resistance
3.1. Bacterial Biocidal Resistance
3.2. Fungal Biocidal Resistance
3.3. Viral Biocidal Resistance
4. Clinical Impact of Antimicrobial Resistance
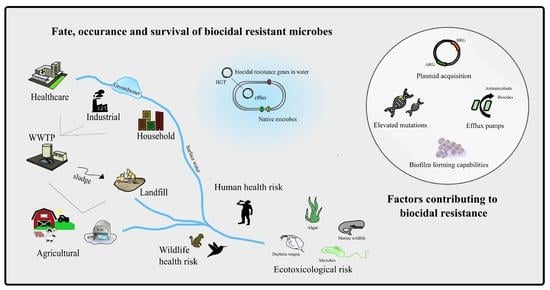
5. Environmental Impact
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lomazzi, M.; Moore, M.; Johnson, A.; Balasegaram, M.; Borisch, B. Antimicrobial resistance—Moving forward? BMC Public Health 2019, 19. [Google Scholar] [CrossRef] [PubMed]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miranda, C.D.; Godoy, F.A.; Lee, M.R. Current Status of the Use of Antibiotics and the Antimicrobial Resistance in the Chilean Salmon Farms. Front. Microbiol. 2018, 9, 1284. [Google Scholar] [CrossRef] [PubMed]
- Van, T.T.H.; Yidana, Z.; Smooker, P.M.; Coloe, P.J. Antibiotic use in food animals worldwide, with a focus on Africa: Pluses and minuses. J. Glob. Antimicrob. Resist. 2020, 20, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Xu, S.; Tang, Z.; Li, Z.; Zhang, L. Use of antimicrobials in food animals and impact of transmission of antimicrobial resistance on humans. Biosaf. Health 2021, 3, 32–38. [Google Scholar] [CrossRef]
- Jasovský, D.; Littmann, J.; Zorzet, A.; Cars, O. Antimicrobial resistance—A threat to the world’s sustainable development. Upsala J. Med. Sci. 2016, 121, 159–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, R.E.; Slayton, R.B.; Stevens, V.W.; Jones, M.M.; Khader, K.; Rubin, M.A.; Jernigan, J.A.; Samore, M.H. Attributable Mortality of Healthcare-Associated Infections Due to Multidrug-Resistant Gram-Negative Bacteria and Methicillin-Resistant Staphylococcus Aureus. Infect. Control. Hosp. Epidemiol. 2017, 38, 848–856. [Google Scholar] [CrossRef]
- Ademe, M.; Girma, F. Candida auris: From Multidrug Resistance to Pan-Resistant Strains. Infect. Drug Resist. 2020, ume 13, 1287–1294. [Google Scholar] [CrossRef]
- Kampf, G.; Todt, D.; Pfaender, S.; Steinmann, E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J. Hosp. Infect. 2020, 104, 246–251. [Google Scholar] [CrossRef] [Green Version]
- Donaghy, J.A.; Jagadeesan, B.; Goodburn, K.; Grunwald, L.; Jensen, O.N.; Jespers, A.D.; Kanagachandran, K.; Lafforgue, H.; Seefelder, W.; Quentin, M.C. Relationship of Sanitizers, Disinfectants, and Cleaning Agents with Antimicrobial Resistance. J. Food Prot. 2019, 82, 889–902. [Google Scholar] [CrossRef]
- Kahrs, R.F. General disinfection guidelines. Rev. Sci. Tech. 1995, 14, 105–163. [Google Scholar] [CrossRef] [Green Version]
- Rutala, W.A.; Weber, D.J. Disinfection and Sterilization in Health Care Facilities: What Clinicians Need to Know. Clin. Infect. Dis. 2004, 39, 702–709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Q.; Lim, J.Y.C.; Xue, K.; Yew, P.Y.M.; Owh, C.; Chee, P.L.; Loh, X.J. Sanitizing agents for virus inactivation and disinfection. View 2020, 1. [Google Scholar] [CrossRef]
- Guan, J.; Chan, M.; Brooks, B.W.; Rohonczy, L. Influence of temperature and organic load on chemical disinfection of Geobacillus steareothermophilus spores, a surrogate for Bacillus anthracis. Can. J. Vet. Res. 2013, 77, 100–104. [Google Scholar] [PubMed]
- Kumar, G.D.; Mishra, A.; Dunn, L.; Townsend, A.; Oguadinma, I.C.; Bright, K.R.; Gerba, C.P. Biocides and Novel Antimicrobial Agents for the Mitigation of Coronaviruses. Front. Microbiol. 2020, 11, 1351. [Google Scholar] [CrossRef]
- Bock, L.J. Bacterial biocide resistance: A new scourge of the infectious disease world? Arch. Dis. Child. 2019, 104, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Wand, M.E. Bacterial Resistance to Hospital Disinfection. In Modeling the Transmission and Prevention of Infectious Disease; Hurst, C.J., Ed.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 19–54. [Google Scholar]
- Maillard, J.Y. Antimicrobial biocides in the healthcare environment: Efficacy, usage, policies, and perceived problems. Ther. Clin. Risk Manag. 2005, 1, 307–320. [Google Scholar] [PubMed]
- Holah, J.T. Cleaning and disinfection practices in food processing. In Hygiene in Food Processing, Principles and Practice; Woodhead Publishing Limited: Cambridge, UK, 2014; pp. 259–304. [Google Scholar]
- Maillard, J.-Y. Resistance of Bacteria to Biocides. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33. [Google Scholar] [CrossRef] [PubMed]
- Vijayakumar, R.; Sandle, T. A review on biocide reduced susceptibility due to plasmid-borne antiseptic-resistant genes—special notes on pharmaceutical environmental isolates. J. Appl. Microbiol. 2019, 126, 1011–1022. [Google Scholar] [CrossRef] [Green Version]
- Mima, T.; Joshi, S.; Gomez-Escalada, M.; Schweizer, H.P. Identification and Characterization of TriABC-OpmH, a Triclosan Efflux Pump of Pseudomonas aeruginosa Requiring Two Membrane Fusion Proteins. J. Bacteriol. 2007, 189, 7600–7609. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Zhao, H.; Han, L.; Shu, W.; Wu, Q.; Ni, Y. Frequency of biocide-resistant genes and susceptibility to chlorhexidine in high-level mupirocin-resistant, methicillin-resistant Staphylococcus aureus (MuH MRSA). Diagn. Microbiol. Infect. Dis. 2015, 82, 278–283. [Google Scholar] [CrossRef] [PubMed]
- Poole, K. Efflux-mediated antimicrobial resistance. J. Antimicrob. Chemother. 2005, 56, 20–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pathania, R.; Sharma, A.; Gupta, V.K. Efflux pump inhibitors for bacterial pathogens: From bench to bedside. Indian J. Med. Res. 2019, 149, 129–145. [Google Scholar] [CrossRef] [PubMed]
- Piddock, L.J.V. Multidrug-resistance efflux pumps—Not just for resistance. Nat. Rev. Microbiol. 2006, 4, 629–636. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G.; Ning, J.; Ahmed, S.; Huang, J.; Ullah, R.; An, B.; Hao, H.; Dai, M.; Huang, L.; Wang, X.; et al. Selection and dissemination of antimicrobial resistance in Agri-food production. Antimicrob. Resist. Infect. Control. 2019, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Pastrana-Carrasco, J.; Garza-Ramos, J.U.; Barrios, H.; Morfin-Otero, R.; Rodríguez-Noriega, E.; Barajas, J.M.; Suárez, S.; Díaz, R.; Miranda, G.; Solórzano, F.; et al. Gene frequency and biocide resistance in extended-spectrum beta-lactamase producing enterobacteriaceae clinical isolates. Rev. Investig. Clin. 2012, 64 Pt 1, 535–540. (In Spanish) [Google Scholar]
- Xiao, T.; Wu, Z.; Shi, Q.; Zhang, X.; Zhou, Y.; Yu, X.; Xiao, Y. A retrospective analysis of risk factors and outcomes in patients with extended-spectrum beta-lactamase-producing Escherichia coli bloodstream infections. J. Glob. Antimicrob. Resist. 2019, 17, 147–156. [Google Scholar] [CrossRef]
- Cadena, M.; Kelman, T.; Marco, M.L.; Pitesky, M. Understanding antimicrobial resistance (AMR) profiles of Salmonella biofilm and planktonic bacteria challenged with disinfectants commonly used during poultry processing. Foods 2019, 87, 275. [Google Scholar] [CrossRef] [Green Version]
- Bjarnsholt, T. The role of bacterial biofilms in chronic infections. APMIS 2013, 121, 1–58. [Google Scholar] [CrossRef]
- Touzel, R.E.; Sutton, J.M.; Wand, M.E. Establishment of a multi-species biofilm model to evaluate chlorhexidine efficacy. J. Hosp. Infect. 2016, 92, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Perumal, P.K.; Wand, M.E.; Sutton, J.M. Evaluation of the effectiveness of the hydrogenperoxide based disinfectants on biofilms formed by Gram-negative pathogens. J. Hosp. Infect. 2014, 87, 227–233. [Google Scholar] [CrossRef]
- Kampf, G. Antibiotic resistance can be enhanced in Gram-positive species by some biocidal agents used for disinfection. Antibiotics 2019, 8, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taheri, N.; Ardebili, A.; Amouzandeh-Nobaveh, A.; Ghaznavi-Rad, E. Frequency of Antiseptic Resistance Among Staphylococcus aureus and Coagulase-Negative Staphylococci Isolated from a University Hospital in Central Iran. Oman Med. J. 2016, 31, 426–432. [Google Scholar] [CrossRef] [PubMed]
- Conceição, T.; Coelho, C.; de Lencastre, H.; Aires-De-Sousa, M. High Prevalence of Biocide Resistance Determinants in Staphylococcus aureus Isolates from Three African Countries. Antimicrob. Agents Chemother. 2015, 60, 678–681. [Google Scholar] [CrossRef] [Green Version]
- Bush, K. Past and Present Perspectives on β-Lactamases. Antimicrob. Agents Chemother. 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slipski, C.J.; Zhanel, G.G.; Bay, D.C. Biocide selective TolC-independent efflux pumps in Enterobacteriaceae. J. Membr. Biol. 2018, 251, 15–33. [Google Scholar] [CrossRef]
- Li, X.-Z.; Plésiat, P.; Nikaido, H. The Challenge of Efflux-Mediated Antibiotic Resistance in Gram-Negative Bacteria. Clin. Microbiol. Rev. 2015, 28, 337–418. [Google Scholar] [CrossRef] [Green Version]
- Romero, J.L.; Grande Burgos, M.J.; Pérez-Pulido, R.; Gálvez, A.; Lucas, R. Resistance to antibiotics, biocides, preservatives and metals in bacteria isolated from seafoods: Co-selection of strains resistant or tolerant to different classes of compounds. Front. Microbiol. 2017, 8, 1650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.J.; Fu, L.; Huang, M.; Zhang, J.P.; Wu, Y.; Zhou, Y.S.; Zeng, J.; Wang, G.X. Frequency of antiseptic resistance genes and reduced susceptibility to biocides in carbapenem-resistant Acinetobacter baumannii. J. Med. Microbiol. 2017, 66, 13–17. [Google Scholar] [CrossRef]
- Su, X.Z.; Chen, J.; Mizushima, T.; Kuroda, T.; Tsuchiya, T. AbeM, an H+-coupled Acinetobacter baumannii multi-drug efflux pump belonging to the MATE family of transporters. Antimicrob. Agents Chemother. 2005, 49, 4362–4364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chitsaz, M.; Booth, L.; Blyth, M.T.; O’Mara, M.L.; Brown, M.H. Multidrug Resistance in Neisseria gonorrhoeae: Identification of Functionally Important Residues in the MtrD Efflux Protein. mBio 2019, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amsalu, A.; Sapula, S.A.; De Barros Lopes, M.; Hart, B.J.; Nguyen, A.H.; Drigo, B.; Turnidge, J.; Leong, L.E.; Venter, H. Efflux pump-driven antibiotic and biocide cross-resistance in Pseudomonas aeruginosa isolated from different ecological niches: A case study in the development of multidrug resistance in environmental hotspots. Microorganisms 2020, 8, 1647. [Google Scholar] [CrossRef]
- Lin, F.; Xu, Y.; Chang, Y.; Liu, C.; Jia, X.; Ling, B. Molecular Characterization of Reduced Susceptibility to Biocides in Clinical Isolates of Acinetobacter baumannii. Front. Microbiol. 2017, 8, 1836. [Google Scholar] [CrossRef]
- Davin-Regli, A. Enterobacter aerogenes and Enterobacter cloacae; Versatile bacterial pathogens confronting antibiotic treatment. Front. Microbiol. 2015, 6, 392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roedel, A.; Vincze, S.; Projahn, M.; Roesler, U.; Robé, C.; Hammerl, J.A.; Noll, M.; Al Dahouk, S.; Dieckmann, R. Genetic but No Phenotypic Associations between Biocide Tolerance and Antibiotic Resistance in Escherichia coli from German Broiler Fattening Farms. Microorganisms 2021, 9, 651. [Google Scholar] [CrossRef] [PubMed]
- Colclough, A.L.; Alav, I.; Whittle, E.E.; Pugh, H.L.; Darby, E.M.; Legood, S.W.; McNeil, H.E.; Blair, J.M. RND efflux pumps in Gram-negative bacteria; regulation, structure and role in antibiotic resistance. Futur. Microbiol. 2020, 15, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Barabote, R.D.; Thekkiniath, J.; Strauss, R.E.; Vediyappan, G.; Fralick, J.A.; Francisco, M.J.S. Xenobiotic Efflux in Bacteria and Fungi: A Genomics Update. Adv. Enzymol. Relat. Areas Mol. Biol. 2011, 77, 237–306. [Google Scholar] [CrossRef] [Green Version]
- Brown, G.D.; Denning, D.W.; Gow, N.A.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4, rv113. [Google Scholar] [CrossRef] [Green Version]
- Meade, E.; Savage, M.; Slattery, M.A.; Garvey, M. Disinfection of Mycotic Species Isolated from Cases of Bovine Mastitis Showing Antifungal Resistance. Cohesive J. Microbiol. Infect. Dis. 2020, 3. [Google Scholar] [CrossRef]
- Meade, E.; Savage, M.; Slattery, M.A.; Garvey, M. An Assessment of Alternative Therapeutic Options for the Treatment of Prolonged Zoonotic Fungal Infections in Companion Animals. J. Microbiol. Biotechnol. 2019, 4, 000149. [Google Scholar]
- Kalem, M.C.; Subbiah, H.; Leipheimer, J.; Glazier, V.E.; Panepinto, J.C. Puf4 medicates post-transcriptional regulation of caspofungin resistance in Cryptococcus neoformans. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Cowen, L.E.; Sanglard, D.; Howard, S.J.; Rogers, P.D.; Perlin, D.S. Mechanisms of Antifungal Drug Resistance. Cold Spring Harb. Perspect. Med. 2015, 5, a019752. [Google Scholar] [CrossRef] [PubMed]
- Eissa, M.E.; El Naby, M.A.; Beshir, M.M. Bacterial vs. fungal spore resistance to peroxygen biocide on inanimate surfaces. Bull. Fac. Pharm. Cairo Univ. 2014, 52, 219–224. [Google Scholar] [CrossRef] [Green Version]
- Rogers, B.D.A.; Kolaczkowski, M.; Carvajal, E.; Balzi, E.; Goffeau, A. The pleitropic drug ABC transporters from Saccharomyces cerevisiae. J. Mol. Microbiol. Biotechnol. 2003, 3, 207–214. [Google Scholar]
- Bhattacharya, S.; Sae-Tia, S.; Fries, B.C. Candidiasis and Mechanisms of Antifungal Resistance. Antibiotics 2020, 9, 312. [Google Scholar] [CrossRef]
- Sandle, T.; Vijayakumar, R.; Al Aboody, M.S.; Saravanakumar, S. In vitro fungicidal activity of biocides against pharmaceutical environmental fungal isolates. J. Appl. Microbiol. 2014, 117, 1267–1273. [Google Scholar] [CrossRef]
- Mihriban, K.; Yasemin, S.; Aycan, Y. The fungicidal efficacy of various commercial disinfectants used in the food industry. Ann. Microbiol. 2006, 56, 325–330. [Google Scholar] [CrossRef]
- Mataraci-Kara, E.; Ataman, M.; Yilmaz, G.; Ozbek-Celik, B. Evaluation of antifungal and disinfectant-resistant Candida species isolated from hospital wastewater. Arch. Microbiol. 2020, 202, 1–8. [Google Scholar] [CrossRef]
- Cadnum, J.L.; Shaikh, A.A.; Piedrahita, C.T.; Sankar, T.; Jencson, A.L.; Larkin, E.L.; Ghannoum, M.A.; Donskey, C.J. Effectiveness of Disinfectants Against Candida auris and other Candida Species. Infect. Control. Hosp. Epidemiol. 2017, 38, 1240–1243. [Google Scholar] [CrossRef] [Green Version]
- Moore, G.; Schelenz, S.; Borman, A.M.; Johnson, E.M.; Brown, C.S. Yeasticidal activity of chemical disinfectants and antiseptics against Candida auris. J. Hosp. Infect. 2017, 97, 371–375. [Google Scholar] [CrossRef] [PubMed]
- Ku, T.S.N.; Walraven, C.J.; Lee, S.A. Candida auris: Disinfectants and Implications for Infection Control. Front. Microbiol. 2018, 9, 726. [Google Scholar] [CrossRef]
- Sisti, M.; Brandi, G.; De Santi, M.; Rinaldi, L.; Schiavano, G.F. Disinfection efficacy of chlorine and peracetic acid alone or in combination against Aspergillus spp. and Candida albicans in drinking water. J. Water Health 2011, 10, 11–19. [Google Scholar] [CrossRef]
- Nuanualsuwan, S.; Cliver, D.O. Capsid Functions of Inactivated Human Picornaviruses and Feline Calicivirus. Appl. Environ. Microbiol. 2003, 69, 350–357. [Google Scholar] [CrossRef] [Green Version]
- Wigginton, K.R.; Pecson, B.M.; Sigstam, T.; Bosshard, F.; Kohn, T. Virus Inactivation Mechanisms: Impact of Disinfectants on Virus Function and Structural Integrity. Environ. Sci. Technol. 2012, 46, 12069–12078. [Google Scholar] [CrossRef]
- Mattle, M.J.; Crouzy, B.; Brennecke, M.; Wigginton, K.R.; Perona, P.; Kohn, T. Impact of Virus Aggregation on Inactivation by Peracetic Acid and Implications for Other Disinfectants. Environ. Sci. Technol. 2011, 45, 7710–7717. [Google Scholar] [CrossRef] [PubMed]
- Weber, D.J.; Rutala, W.A.; Healthcare Infection Control Practices Advisory Committee (HICPAC). Guideline for Disinfection and Sterilization in Healthcare Facilities. 2019. Available online: https://www.cdc.gov/infectioncontrol/guidelines/disinfection/index.html (accessed on 22 March 2021).
- Ansaldi, F.; Banfi, F.; Morelli, P.; Valle, L.; Durando, P.; Sticchi, L.; Contos, S.; Gasparini, R.; Crovari, P. SARS-CoV, influenza A and syncitial respiratory virus resistance against common disinfectants and ultraviolet irradiation. J. Prev. Med. Hyg. 2004, 45, 5–8. [Google Scholar]
- Piret, J.; Roy, S.; Gagnon, M.; Landry, S.; Désormeaux, A.; Omar, R.F.; Bergeron, M.G. Comparative Study of Mechanisms of Herpes Simplex Virus Inactivation by Sodium Lauryl Sulfate and n-Lauroylsarcosine. Antimicrob. Agents Chemother. 2002, 46, 2933–2942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Sayah, M.H. Chemical disinfectants of COVID-19: An overview. J. Water Health 2020, 18, 843–848. [Google Scholar] [CrossRef]
- Wang, X.-W.; Li, J.-S.; Jin, M.; Zhen, B.; Kong, Q.-X.; Song, N.; Xiao, W.-J.; Yin, J.; Wei, W.; Wang, G.-J.; et al. Study on the resistance of severe acute respiratory syndrome-associated coronavirus. J. Virol. Methods 2005, 126, 171–177. [Google Scholar] [CrossRef]
- McDonnell, G.; Russell, A.D. Antiseptics and Disinfectants: Activity, Action, and Resistance. Clin. Microbiol. Rev. 1999, 12, 147–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bosco-Borgeat, M.E.; Mazza, M.; Taverna, C.G.; Córdoba, S.; Murisengo, O.A.; Vivot, W.; Davel, G. Amino acid substitution in Cryptococcus neoformans lanosterol 14-α-demethylase involved in fluconazole resistance in clinical isolates. Rev. Argent. Microbiol. 2016, 48, 137–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carolus, H.; Pierson, S.; Lagrou, K.; Van Dijck, P. Amphotericin B and Other Polyenes—Discovery, Clinical Use, Mode of Action and Drug Resistance. J. Fungi 2020, 6, 321. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, M.; Khaithir, T.M.N.; Santhanam, J. Determination of azole antifungal drug resistance mechanisms involving Cyp51A gene in clinical isolates of Aspergillus fumigatus and Aspergillus niger. Malays. J. Microbiol. 2016, 12, 205–210. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. HIV Drug Resistance Report 2019; Licence CC BY-NC-SA 3.0 IGO; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Hao, H.; Cheng, G.; Iqbal, Z.; Ai, X.; Hussain, H.I.; Huang, L.; Dai, M.; Wang, Y.; Liu, Z.; Yuan, Z. Benefits and risks of antimicrobial use in food-producing animals. Front. Microbiol. 2014, 5, 288. [Google Scholar] [CrossRef] [Green Version]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef]
- Kampf, G. Biocidal agents used for disinfection can enhance antibiotic resistance in gram-negative species. Antibiotics 2018, 7, 110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehrad, B.; Clark, N.M.; Zhanel, G.G.; Lynch, J.P. Antimicrobial Resistance in Hospital-Acquired Gram-Negative Bacterial Infections. Chest 2015, 147, 1413–1421. [Google Scholar] [CrossRef] [Green Version]
- Kang, C.; Kim, S.; Kim, H.; Park, S.; Choe, Y.; Oh, M.; Kim, E.; Choe, K. Pseudomonas aeruginosa Bacteremia: Risk Factors for Mortality and Influence of Delayed Receipt of Effective Antimicrobial Therapy on Clinical Outcome. Clin. Infect. Dis. 2003, 37, 745–751. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, A.; Nsonwu, O.; Johnson, A.; Hope, R. Estimating the incidence and 30-day all-cause mortality rate of Escherichia coli bacteraemia in England by 2020/21. J. Hosp. Infect. 2018, 98, 228–231. [Google Scholar] [CrossRef]
- Karatzas, K.A.G.; Webber, M.A.; Jorgensen, F.; Woodward, M.J.; Piddock, L.J.V.; Humphrey, T.J. Prolonged treatment of Salmonella enterica serovar Typhimurium with commercial disinfectants selects for multiple antibiotic resistance, increased efflux and reduced invasiveness. J. Antimicrob. Chemother. 2007, 60, 947–955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bougnoux, M.-E.; Brun, S.; Zahar, J.-R. Healthcare-associated fungal outbreaks: New and uncommon species, New molecular tools for investigation and prevention. Antimicrob. Resist. Infect. Control. 2018, 7, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seyedmousavi, S.; Guillot, J.; Tolooe, A.; Verweij, P.; De Hoog, G. Neglected fungal zoonoses: Hidden threats to man and animals. Clin. Microbiol. Infect. 2015, 21, 416–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chow, E.J.; Mermel, L.A. Hospital-Acquired Respiratory Viral Infections: Incidence, Morbidity, and Mortality in Pediatric and Adult Patients. Open Forum Infect. Dis. 2017, 4, ofx006. [Google Scholar] [CrossRef] [Green Version]
- Micek, S.T.; Chew, B.; Hampton, N.; Kollef, M.H. A Case-Control Study Assessing the Impact of Nonventilated Hospital-Acquired Pneumonia on Patient Outcomes. Chest 2016, 150, 1008–1014. [Google Scholar] [CrossRef] [PubMed]
- Dumas, O.; Varraso, R.; Boggs, K.M.; Quinot, C.; Zock, J.-P.; Henneberger, P.K.; Speizer, F.E.; Le Moual, N.; Camargo, C.A., Jr. Association of occupational exposure to disinfectants with incidence of chronic obstructive pulmonary disease among US female nurses. JAMA Netw. Open 2019, 2, e1913563. [Google Scholar] [CrossRef]
- Nabi, G.; Wang, Y.; Hao, Y.; Khan, S.; Wu, Y.; Li, D. Massive use of disinfectants against COVID-19 poses potential risks to urban wildlife. Environ. Res. 2020, 188, 109916. [Google Scholar] [CrossRef]
- Xie, H.; Du, J.; Chen, J. Concerted Efforts Are Needed to Control and Mitigate Antibiotic Pollution in Coastal Waters of China. Antibiotics 2020, 9, 88. [Google Scholar] [CrossRef] [Green Version]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules 2018, 23, 795. [Google Scholar] [CrossRef] [Green Version]
- Kraemer, S.A.; Ramachandran, A.; Perron, G.G. Antibiotic Pollution in the Environment: From Microbial Ecology to Public Policy. Microorganisms 2019, 7, 180. [Google Scholar] [CrossRef] [Green Version]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hora, P.I.; Pati, S.G.; McNamara, P.J.; Arnold, W.A. Increased Use of Quaternary Ammonium Compounds during the SARS-CoV-2 Pandemic and Beyond: Consideration of Environmental Implications. Environ. Sci. Technol. Lett. 2020, 7, 00437. [Google Scholar] [CrossRef]
- Zhang, C.; Cui, F.; Zeng, G.-M.; Jiang, M.; Yang, Z.-Z.; Yu, Z.-G.; Zhu, M.-Y.; Shen, L.-Q. Quaternary ammonium compounds (QACs): A review on occurrence, fate and toxicity in the environment. Sci. Total. Environ. 2015, 518–519, 352–362. [Google Scholar] [CrossRef] [PubMed]
| Enzyme Type | Representative Enzymes | Known Substrates | Inhibitor Profile | Clinically Associated Pathogens | Biocidal Resistance | |
|---|---|---|---|---|---|---|
| Serine β-lactamases | Penicillinase | PC1/blaZ | Penicillins | CA and TZ | MRSA | qacA/B (acquired), norA and lmrS (intrinsic) genes encoding MFS pumps. MecA (MATE superfamily) and sepA multidrug efflux pump genes. SMR pumps encoded by smr (also known as qacC/D and Ebr), qacG, qacH and qacEΔ1 (acquired) [35,36,37] |
| Broad- spectrum (TEM, SHV-type) | TEM-1, -2 and -13, SHV-1 and -11 | Penicillins and 1st-generation cephalosporins [38] | CA, TZ and SB | Enterobacteriaceae (E. coli, K. pneumonia, Proteus sp.) non fermenters (i.e., Pseudomonas aeruginosa., Acinetobacter baumannii) and Neisseria gonorrhoeae | Acquired efflux resistance to QACs and chlorhexidine encoded by qacEΔ1, qacE, qacG, qacH and emrE (SMR), qacA (MFS) and cep A genes common in many Enterobacteriaceae [39,40] and non-fermenters [41,42] Multidrug efflux MATE pumps (chromosomally encoded) conferring resistance to biocides and antimicrobials, examples include YdhE of E. coli, PmpM of P. aeruginosa, and AbeM of A. baumannii [43] Upregulation of chromosomally encoded RND pumps conferring cross-resistance to biocides, antimicrobials and other agents (dyes, metals), examples include AcrAB-TolC, AcrEF-TolC in E. coli and other Enterobacteriaceae [39] MtrD in N. gonorrhoeae [44] MexAB-OprM, MexCD-OprJ, MexEF-OprN and MexJK pumps in Pseudomonas [45] AdeABC, AdeFGH, AdeIJK and AbeD efflux systems in A. baumannii [46] | |
| TEM-30 and -31, SHV-10 | Penicillins | Reduced binding to CA or inhibitor resistant apart from AV | ||||
| ESBL (TEM, SHV, PER, VEB, CTX-M-type) | TEM-3, and -10, SHV-3, CTX-M-1, -14, -15 and -44, PER-1, VEB-1 | Penicillins, 1st, 2nd- and 3rd-generation cephalosporins and monobactam | CA, TZ, SB and AV | |||
| TEM-50 and -158 | Reduced binding to CA or inhibitor resistant apart from AV | |||||
| Carbenicillinase | PESE-1, -3 and -4, CARB-1 | Penicillins and carbenicillin | CA, TZ and SB | |||
| Carbapenemase (KPC, GES, SME-type) | KPC-2 and -10, IMI-1, SME-1, and -2, GES-2 and -7 | All beta lactams | Variable to CA, TZ and AV | P. aeruginosa, and K. pneumonia (and other Enterobacteriaceae) | ||
| OXA-type (Broad spectrum, ESBL and Carbapenemase) | OXA-1, OXA-9, OXA-10, OXA-2 [38] | Penicillins (oxacillin, cloxacillin) | Variable to CA, TZ and AV | Enterobacteriaceae (K. pneumonia, E. coli, Enterobacter sp.), nonfermenters and Neisseria gonorrhoeae | ||
| OXA-11, OXA-14, OXA-15, | Penicillins, 3rd-generation cephalosporins, monobactams | |||||
| OXA-3, OXA-51, OXA-58, OXA-23, OXA-48 | All beta lactams/carbapenems | |||||
| AmpC cephamycinases | AmpC (chromosomal encoded) | All beta lactams except carbapenems | Inhibitor resistant apart from AV | Citrobacter, Serratia, Enterobacter spp., and P. aeruginosa (expression usually inducible) and Enterobacteriaceae (not as inducible) | Studies report on the presence of efflux pumps belonging to the MATE and RND families in Enterobacter, where AmpC is inducible in these species [47] qacE∆1 is commonly reported in enteric pathogens, being associated with class 1 integrons that carry multiple gene cassettes including AmpC β-lactamases [48] | |
| MOX, ACC, FOX, DHA, CMY, MIR-type (plasmid encoded) | Non fermenters and Enterobacteriaceae | |||||
| Metallo-β-lactamases | Carbapenemases (IMP, VIM, NDM-type) | IMP-1, VIM -1 and -2, NDM-1 [38] | All beta lactams except aztreonam | EDTA or 1-10 phenanthroline, mercaptopropionic acid or sodium mercaptoacetic acid and dipicolinic acid | Pseudomonas and Acinetobacter sp. | RND efflux pumps on plasmids that carry resistance determinants such as blaNDM-1 have been reported [41,49] Association of qac genes with the presence of NDM, VIM and IMP beta lactamases reported in clinical A. baumanii [42] |
| Medically Important Pathogen | Associated Disease | Antimicrobial Resistance | Biocidal Resistance | |
|---|---|---|---|---|
| Fungal | Candida albicans | Candidemia, mucosal candidiasis, cutaneous infections | Mutations in ERG11 and Upc2p, and overexpression of Cdr1, Cdr2 and Mdr1 confer azole resistance Polyene resistance is linked to changes in ERG3 and ERG6 Mutations in CaFKS1 confer resistance to echinocandins [58] | Fungal biocide resistance is not yet completely understood, being related to multiple defence mechanisms, including mutations, inducible efflux, exclusion or reduced access of antiseptic or disinfectant (chlorhexidine), enzymatic inactivation (formaldehyde) and phenotypic modulation (alcohol) [59,74] Virulence factors such as biofilm-forming capabilities and melanin further contribute to protection against biocides in fungi |
| Cryptococcus neoformans | Cryptococcal meningitis, pulmonary cryptococcosis, cutaneous infections | Mutations in ERG11, overexpression of ERG11 due to chromosome 1 duplication and upregulation of AFR1 gene (encodes ABC transporter) confer resistance to azoles [75] Mutation in ERG2 resulting in its inactivation, confers resistance to amphotericin b [76] | ||
| Aspergillus niger | Pulmonary aspergillosis, Aspergillus bronchitis, allergic bronchopulmonary aspergillosis (ABPA) | Azole resistance related to point mutations in Cyp51A gene, overexpression of Cyp51A gene and upregulation of efflux pumps [77] | ||
| Viral | Human papillomavirus (HPV) (nonenveloped) | Cervical cancer | No treatment available | Nonenveloped viruses are more resistant to biocides, showing reduced susceptibility/resistance to lipophilic agents such as Qacs [13] |
| Human immunodeficiency virus (HIV) (enveloped) | Acquired immunodeficiency syndrome (AIDS) | Drug resistance is caused by changes in the genetic structure of HIV that affect the ability of drugs (e.g., HAART) to block the replication of the virus [78] | Enveloped viruses are the least resistant to inactivation by biocides, where their lipid envelope is easily compromised by most disinfectants and antiseptics [13] | |
| Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (enveloped) | Respiratory illness | No treatment available |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meade, E.; Slattery, M.A.; Garvey, M. Biocidal Resistance in Clinically Relevant Microbial Species: A Major Public Health Risk. Pathogens 2021, 10, 598. https://doi.org/10.3390/pathogens10050598
Meade E, Slattery MA, Garvey M. Biocidal Resistance in Clinically Relevant Microbial Species: A Major Public Health Risk. Pathogens. 2021; 10(5):598. https://doi.org/10.3390/pathogens10050598
Chicago/Turabian StyleMeade, Elaine, Mark Anthony Slattery, and Mary Garvey. 2021. "Biocidal Resistance in Clinically Relevant Microbial Species: A Major Public Health Risk" Pathogens 10, no. 5: 598. https://doi.org/10.3390/pathogens10050598