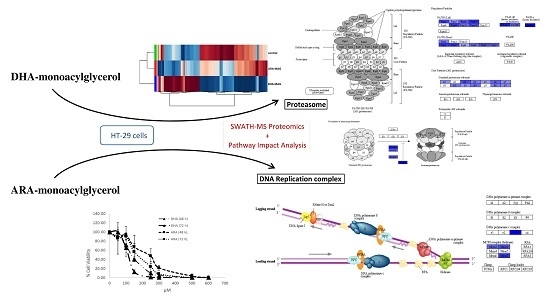
SWATH Differential Abundance Proteomics and Cellular Assays Show In Vitro Anticancer Activity of Arachidonic Acid- and Docosahexaenoic Acid-Based Monoacylglycerols in HT-29 Colorectal Cancer Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Oil Samples and Purification of MAG
2.2. Cell Assays
2.3. SWATH-MS Differential Abundance Proteomics Analysis
2.4. Pathway and Gene Ontology (GO) Analysis
2.5. Validation by Parallel Reaction Monitoring (PRM) Analysis
2.6. Statistical Analysis
3. Results
3.1. DHA- and ARA-MAG Showed Differential and Concentration-Dependent Effects on HT-29 Cell Viability, Cell Membrane Integrity, and Apoptosis
3.2. SWATH Quantitation of 1882 proteins Showed That DHA- and ARA-MAG Differentially Affect the Whole Proteome of HT-29 Cells
3.3. Pathway and GO Analysis Showed Different Mechanisms of Action of DHA- and ARA-MAG on HT-29 cells
3.4. SWATH Proteomics Analysis was validated by PRM
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Castro González, M.I. Ácidos grasos omega 3: Beneficios y fuentes. Interciencia 2002, 27, 128–136. [Google Scholar]
- Ramírez, M.; Amate, L.; Gil, A. Absorption and distribution of dietary fatty acids from different sources. Early Hum. Dev. 2001, 65, S95–S101. [Google Scholar] [CrossRef]
- Philippoussis, F.; Przybytkowski, E.; Fortin, M.; Arguin, C.; Pande, S.V.; Steff, A.M.; Hugo, P. Derivatives of monoglycerides as apoptotic agents in T-cells. Cell Death Differ. 2001, 8, 1103–1112. [Google Scholar] [CrossRef] [PubMed]
- Rincón-Cervera, M.A.; Rodríguez-García, I.; Guil-Guerrero, J.L. Purification of GLA-triglycerides from evening primrose oil by gravimetric column chromatography. JAOCS J. Am. Oil Chem. Soc. 2009, 86, 605–609. [Google Scholar] [CrossRef]
- Ho, S.; Storch, J. Common mechanisms of monoacylglycerol and fatty acid uptake by human intestinal Caco-2 cells. Am. J. Physiol. Cell Physiol. 2009, 281, 1106–1117. [Google Scholar] [CrossRef]
- Guil-Guerrero, J.L.; Belarbi, E.-H.; Rebolloso-Fuentes, M.M. Eicosapentaenoic and arachidonic acids purification from the red microalga Porphyridium cruentum. Bioseparation 2000, 9, 299–306. [Google Scholar] [CrossRef]
- Stillwell, W.; Wassall, S.R. Docosahexaenoic acid: Membrane properties of a unique fatty acid. Chem. Phys. Lipids 2003, 126, 1–27. [Google Scholar] [CrossRef]
- Sijtsma, L.; de Swaaf, M.E. Biotechnological production and applications of the ω-3 polyunsaturated fatty acid docosahexaenoic acid. Appl. Microbiol. Biotechnol. 2004, 64, 146–153. [Google Scholar] [CrossRef]
- Joint FAO/WHO Expert Consultation. Fats and Fatty Acids in Human Nutrition; Report of an expert consultation; FAO Food and Nutrition Paper: Rome, Italy, 2010; Volume 91, pp. 1–116. [Google Scholar]
- Rodríguez-Cruz, M.; Tovar, A.R.; del Prado, M.; Torres, N. Mecanismos moleculares de acción de los ácidos grasos poliinsaturados y sus beneficios en la salud. Rev. Invest. Clin. 2005, 57, 457–472. [Google Scholar]
- Kang, K.S.; Wang, P.; Yamabe, N.; Fukui, M.; Jay, T.; Zhu, B.T. Docosahexaenoic Acid Induces Apoptosis in MCF-7 Cells in Vitro and in Vivo via Reactive Oxygen Species Formation and Caspase 8 Activation. PLoS ONE 2010, 5, e10296. [Google Scholar] [CrossRef] [Green Version]
- Pompeia, C.; Freitas, J.J.S.; Kim, J.S.; Zyngier, S.B.; Curi, R. Arachidonic acid cytotoxicity in leukocytes: Implications of oxidative stress and eicosanoid synthesis. Biol. Cell 2002, 94, 251–265. [Google Scholar] [CrossRef]
- Morin, C.; Fortin, S.; Cantin, A.; Sirois, M.; Sirois, C.; Rizcallah, E.; Rousseau, E. Anti-Cancer Effects of a New Docosahexaenoic Acid Monoacylglyceride in Lung Adenocarcinoma. Recent Pat. Anticancer. Drug Discov. 2013, 8, 319–334. [Google Scholar] [CrossRef] [PubMed]
- Morin, C.; Rousseau, É.; Fortin, S. Anti-proliferative effects of a new docosapentaenoic acid monoacylglyceride in colorectal carcinoma cells. Prostaglandins Leukot. Essent. Fat. Acids 2013, 89, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.X.; Wasan, K.M. Effects of Monoglycerides on Rhodamine 123 Accumulation, Estradiol 17 β-D-Glucuronide Bidirectional Transport and MRP2 Protein Expression within Caco-2 cells. J. Pharm. Pharm. Sci. 2008, 11, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulthess, G.; Lipka, G.; Compassi, S.; Boffelli, D.; Weber, F.E.; Paltauf, F.; Hauser, H. Absorption of Monoacylglycerols by Small Intestinal Brush Border Membrane. Biochemistry 1994, 33, 4500–4508. [Google Scholar] [CrossRef]
- Lewin, T.M.; Kim, J.-H.; Granger, D.A.; Vance, J.E.; Coleman, R.A. Acyl-CoA Synthetase Isoforms 1, 4, and 5 Are Present in Different Subcellular Membranes in Rat Liver and Can Be Inhibited Independently. J. Biol. Chem. 2001, 276, 24674–24679. [Google Scholar] [CrossRef] [Green Version]
- Mashek, D.G.; Coleman, R.A. Cellular fatty acid uptake: The contribution of metabolism. Curr. Opin. Lipidol. 2006, 17, 274–278. [Google Scholar] [CrossRef]
- Poursharifi, P.; Madiraju, S.R.M.; Prentki, M. Monoacylglycerol signalling and ABHD6 in health and disease. Diabetes Obes. Metab. 2017, 19, 76–89. [Google Scholar] [CrossRef] [Green Version]
- Gajda, A.M.; Storch, J. Enterocyte fatty acid-binding proteins (FABPs): Different functions of liver and intestinal FABPs in the intestine. Prostaglandins Leukot. Essent. Fat. Acids 2015, 93, 9–16. [Google Scholar] [CrossRef] [Green Version]
- Ortea, I.; Gonzalez-Fernandez, M.J.; Ramos-Bueno, R.P.; Guil Guerrero, J.L. Proteomics Study Reveals That Docosahexaenoic and Arachidonic Acids Exert Different in Vitro Anticancer Activities in Colorectal Cancer Cells. J. Agric. Food Chem. 2018, 66, 6003–6012. [Google Scholar] [CrossRef]
- González-Fernández, M.J.; Ramos-Bueno, R.P.; Rodríguez-García, I.; Guil-Guerrero, J.L. Purification process for MUFA-and PUFA-based monoacylglycerols from edible oils. Biochimie 2017, 139, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Ruiz, J.; Belarbi, E.-H.; Sánchez, J.L.G.; Alonso, D.L. Rapid simultaneous lipid extraction and transesterification for fatty acid analyses. Biotechnol. Tech. 1998, 12, 689–691. [Google Scholar] [CrossRef]
- Ramos-Bueno, R.P.; González-Fernández, M.J.; Guil-Guerrero, J.L. Various Acylglycerols from Common Oils Exert Different Antitumor Activities on Colorectal Cancer Cells. Nutr. Cancer 2016, 68, 518–529. [Google Scholar] [CrossRef] [PubMed]
- Maclean, B.; Tomazela, D.M.; Shulman, N.; Chambers, N.; Finney, G.L.; Frewen, B.; Kern, R.; Tabb, D.; Liebler, D.C.; MacCoss, M.J. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Storey, J.D.; Bass, A.J.; Dabney, A.; Robinson, D. Qvalue: Q-Value Estimation for False Discovery Rate Control. R Package Version 2.10.0. Available online: http://github.com/jdstorey/qvalue (accessed on 18 April 2018).
- Lyubachevskaya, G.; Boyle-Roden, E. Kinetics of 2-monoacylglycerol acyl migration in model chylomicra. Lipids 2000, 35, 1353–1358. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; Fu, T.; Wang, L.U.; Lai, Y.; Wang, Y.; Xu, C.; Huang, L.; Guo, Y. Role of autophagy in the ω-3 long chain polyunsaturated fatty acid-induced death of lung cancer A549 cells. Oncol. Lett. 2015, 9, 2736–2742. [Google Scholar] [CrossRef] [Green Version]
- Scorrano, L.; Penzo, D.; Petronilli, V.; Pagano, F.; Bernardi, P. Arachidonic acid causes cell death through the mitochondrial permeability transition. Implications for tumor necrosis factor-alpha aopototic signaling. J. Biol. Chem. 2000, 276, 12035–12040. [Google Scholar] [CrossRef] [Green Version]
- Wiȩckowski, M.R.; Brdiczka, D.; Wojtczak, L. Long-chain fatty acids promote opening of the reconstituted mitochondrial permeability transition pore. FEBS Lett. 2000, 484, 61–64. [Google Scholar] [CrossRef] [Green Version]
- Fotakis, G.; Timbrell, J.A. In vitro cytotoxicity assays: Comparison of LDH, neutral red, MTT and protein assay in hepatoma cell lines following exposure to cadmium chloride. Toxicol. Lett. 2006, 160, 171–177. [Google Scholar] [CrossRef]
- Konishi, T.; Satsu, H.; Hatsugai, Y.; Aizawa, K.; Inakuma, T.; Nagata, S.; Sakuda, S.; Nagasawa, H.; Shimizu, M. Inhibitory effect of a bitter melon extract on the P-glycoprotein activity in intestinal Caco-2 cells. Br. J. Pharmacol. 2004, 143, 379–387. [Google Scholar] [CrossRef] [Green Version]
- Barta, C.A.; Sachs-Barrable, K.; Feng, F.; Wasan, K.M. Effects of Monoglycerides on P-Glycoprotein: Modulation of the Activity and Expression in Caco-2 Cell Monolayers. Mol. Pharm. 2008, 5, 863–875. [Google Scholar] [CrossRef] [PubMed]
- Trombetta, A.; Maggiora, M.; Martinasso, G.; Cotogni, P.; Canuto, R.A.; Muzio, G. Arachidonic and docosahexaenoic acids reduce the growth of A549 human lung-tumor cells increasing lipid peroxidation and PPARs. Chem. Biol. Interact. 2007, 165, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Schwenk, R.W.; Holloway, G.P.; Luiken, J.J.F.P.; Bonen, A.; Glatz, J.F.C. Fatty acid transport across the cell membrane: Regulation by fatty acid transporters. Prostaglandins Leukot. Essent. Fat. Acids 2010, 82, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Dommels, Y.E.M.; Haring, M.M.G.; Keestra, N.G.M.; Alink, G.M.; van Bladeren, P.J.; van Ommen, B. The role of cyclooxygenase in n-6 and n-3 polyunsaturated fatty acid mediated effects on cell proliferation, PGE2 synthesis and cytotoxicity in human colorectal carcinoma cell lines. Carcinogenesis 2003, 24, 385–392. [Google Scholar] [CrossRef] [Green Version]
- Catalani, S.; Carbonaro, V.; Palma, F.; Arshakyan, M.; Galati, R.; Nuvoli, B.; Battistelli, S.; Canestrari, F.; Benedetti, S. Metabolism modifications and apoptosis induction after CellfoodTM administration to leukemia cell lines. J. Exp. Clin. Cancer Res. 2013, 32, 63. [Google Scholar] [CrossRef] [Green Version]
- Kato, T.; Kolenic, N.; Pardini, R.S. Docosahexaenoic Acid (DHA), a Primary Tumor Suppressive Omega-3 Fatty Acid, Inhibits Growth of Colorectal Cancer Independent of p53 Mutational Status. Nutr. Cancer 2007, 58, 178–187. [Google Scholar] [CrossRef]
- Abdi, J.; Garssen, J.; Faber, J.; Redegeld, F.A. Omega-3 fatty acids, EPA and DHA induce apoptosis and enhance drug sensitivity in multiple myeloma cells but not in normal peripheral mononuclear cells. J. Nutr. Biochem. 2014, 25, 1254–1262. [Google Scholar] [CrossRef]
- Gillet, L.C.; Navarro, P.; Tate, S.; Röst, H.; Selevsek, N.; Reiter, L.; Aebersold, R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: A new concept for consistent and accurate proteome analysis. Mol. Cell. Proteom. 2012, 11, O111-016717. [Google Scholar] [CrossRef] [Green Version]
- Selevsek, N.; Chang, C.-Y.; Gillet, L.C.; Navarro, P.; Bernhardt, O.M.; Reiter, L.; Cheng, L.-Y.; Vitek, O.; Aebersold, R. Reproducible and Consistent Quantification of the Saccharomyces cerevisiae Proteome by SWATH-mass spectrometry. Mol. Cell. Proteom. 2015, 14, 739–749. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Bilbao, A.; Bruderer, T.; Luban, J.; Strambio-De-Castillia, C.; Lisacek, F.; Hopfgartner, G.; Varesio, E. The Use of Variable Q1 Isolation Windows Improves Selectivity in LC–SWATH–MS Acquisition. J. Proteome Res. 2015, 14, 4359–4371. [Google Scholar] [CrossRef]
- Liu, B.; Qu, L.; Yan, S. Cyclooxygenase-2 promotes tumor growth and suppresses tumor immunity. Cancer Cell Int. 2015, 15, 106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greenhough, A.; Smartt, H.; Moore, A.; Roberts, H.; Williams, A.; Paraskeva, C.; Kaidi, A. The COX-2/PGE2 pathway: Key roles in the hallmarks of cancer and adaptation to the tumour microenvironment. Carcinogenesis 2009, 30, 377–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarca, A.L.; Draghici, S.; Khatri, P.; Hassan, S.S.; Mittal, P.; Kim, J.-S.; Kim, C.J.; Kusanovic, J.P.; Romero, R. A novel signaling pathway impact analysis. Bioinformatics 2009, 25, 75–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, G.M.; Falke, S.; Goldberg, A.L.; Slaughter, C.A.; DeMartino, G.N.; Gogol, E.P. Structural and functional effects of PA700 and modulator protein on proteasomes. J. Mol. Biol. 1997, 273, 646–657. [Google Scholar] [CrossRef] [PubMed]
- Rechsteiner, M.; Hill, C.P. Mobilizing the proteolytic machine: Cell biological roles of proteasome activators and inhibitors. Trends Cell Biol. 2005, 15, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.; Elliott, P.J. New agents in cancer clinical trials. Oncogene 2000, 19, 6687–6692. [Google Scholar] [CrossRef] [Green Version]
- Kisselev, A.F.; Goldberg, A.L. Proteasome inhibitors: From research tools to drug candidates. Chem. Biol. 2001, 8, 739–758. [Google Scholar] [CrossRef] [Green Version]
- Stillman, B. Smart machines at the DNA replication fork. Cell 1994, 78, 725–728. [Google Scholar] [CrossRef]
- Peterson, A.C.; Russell, J.D.; Bailey, D.J.; Westphall, M.S.; Coon, J.J. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol. Cell. Proteom. 2012, 11, 1475–1488. [Google Scholar] [CrossRef] [Green Version]
- Bourmaud, A.; Gallien, S. Parallel reaction monitoring using quadrupole-Orbitrap mass spectrometer: Principle and applications. Proteomics 2016, 16, 2146–2159. [Google Scholar] [CrossRef]
- Castellone, M.D.; Teramoto, H.; Gutkind, J.S. Cyclooxygenase-2 and Colorectal Cancer Chemoprevention: The β-Catenin Connection. Cancer Res. 2006, 66, 11085–11088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreira, L.; Castells, A. Cyclooxygenase as a target for colorectal cancer chemoprevention. Curr. Drug Targets 2011, 12, 1888–1894. [Google Scholar] [CrossRef] [PubMed]
- Stefan, N.; Fritsche, A.; Häring, H.; Stumvoll, M. Acute stimulation of leptin concentrations in humans during hyperglycemic hyperinsulinemia. Influence of free fatty acids and fasting. Int. J. Obes. 2001, 25, 138. [Google Scholar]






| Protein ID | Protein Name | ARA-MAG to Control | |||||
| SWATH | PRM Validation | ||||||
| FC | q-Value | Significant Change | FC | adj. p-Value | Significant Change | ||
| MCM2 | DNA replication licensing factor MCM2 | 0.46 | 8.3 × 10−0 | ↓ | 0.56 | 2.1 × 10−2 | ↓ |
| MCM7 | DNA replication licensing factor MCM7 | 0.37 | 6.4 × 10−4 | ↓ | 0.57 | 2.9 × 10−2 | ↓ |
| PSMF1 | Proteasome inhibitor PI31 subunit | 0.44 | 7.4 × 10−3 | ↓ | 1.00 | 1.0 × 10+0 | no |
| PSME3 | Proteasome activator complex subunit 3 | 0.47 | 1.3 × 10−4 | ↓ | 0.81 | 3.7 × 10−2 | ↓ |
| PSA3 | Proteasome subunit alpha type-3 | 1.05 | 3.6 × 10−1 | no | 1.18 | 2.2 × 10−1 | no |
| ABHD2 | Abhydrolase domain-containing protein 2 | 6.14 | 2.4 × 10−6 | ↑ | 5.81 | 0 | ↑ |
| SRXN1 | Sulfiredoxin-1 | 4.81 | 1.3 × 10−4 | ↑ | 5.12 | 0 | ↑ |
| TEBP | Prostaglandin E synthase 3 | 0.97 | 3.6 × 10−1 | no | 1.03 | 8.7 × 10−1 | no |
| AIFM1 | Apoptosis-inducing factor 1, mitochondrial | 1.24 | 6.7 × 10−2 | no | 1.42 | 1.3 × 10−1 | no |
| Protein ID | Protein Name | DHA-MAG to Control | |||||
| SWATH | PRM Validation | ||||||
| FC | q-Value | Significant Change | FC | adj. p-Value | Significant Change | ||
| MCM2 | DNA replication licensing factor MCM2 | 0.29 | 4.8 × 10−6 | ↓ | 0.24 | 0 | ↓ |
| MCM7 | DNA replication licensing factor MCM7 | 0.21 | 2.6 × 10−6 | ↓ | 0.16 | 0 | ↓ |
| PSMF1 | Proteasome inhibitor PI31 subunit | 0.13 | 1.8 × 10−5 | ↓ | 0.50 | 2.1 × 10−2 | ↓ |
| PSME3 | Proteasome activator complex subunit 3 | 0.13 | 1.4 × 10−7 | ↓ | 0.24 | 0 | ↓ |
| PSA3 | Proteasome subunit alpha type-3 | 0.40 | 6.7 × 10−7 | ↓ | 0.31 | 0 | ↓ |
| ABHD2 | Abhydrolase domain-containing protein 2 | 1.35 | 1.0 × 10−1 | no | 0.97 | 8.4 × 10−1 | no |
| SRXN1 | Sulfiredoxin-1 | 0.40 | 9.7 × 10−5 | ↓ | 0.38 | 7.0 × 10−4 | ↓ |
| TEBP | Prostaglandin E synthase 3 | 0.14 | 2.0 × 10−7 | ↓ | 0.08 | 0 | ↓ |
| AIFM1 | Apoptosis-inducing factor 1, mitochondrial | 2.34 | 1.6 × 10−6 | ↑ | 1.82 | 1.1 × 10−3 | ↑ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
González-Fernández, M.J.; Fabrikov, D.; Ramos-Bueno, R.P.; Guil-Guerrero, J.L.; Ortea, I. SWATH Differential Abundance Proteomics and Cellular Assays Show In Vitro Anticancer Activity of Arachidonic Acid- and Docosahexaenoic Acid-Based Monoacylglycerols in HT-29 Colorectal Cancer Cells. Nutrients 2019, 11, 2984. https://doi.org/10.3390/nu11122984
González-Fernández MJ, Fabrikov D, Ramos-Bueno RP, Guil-Guerrero JL, Ortea I. SWATH Differential Abundance Proteomics and Cellular Assays Show In Vitro Anticancer Activity of Arachidonic Acid- and Docosahexaenoic Acid-Based Monoacylglycerols in HT-29 Colorectal Cancer Cells. Nutrients. 2019; 11(12):2984. https://doi.org/10.3390/nu11122984
Chicago/Turabian StyleGonzález-Fernández, María José, Dmitri Fabrikov, Rebeca P. Ramos-Bueno, José Luis Guil-Guerrero, and Ignacio Ortea. 2019. "SWATH Differential Abundance Proteomics and Cellular Assays Show In Vitro Anticancer Activity of Arachidonic Acid- and Docosahexaenoic Acid-Based Monoacylglycerols in HT-29 Colorectal Cancer Cells" Nutrients 11, no. 12: 2984. https://doi.org/10.3390/nu11122984