A Gemini Cationic Lipid with Histidine Residues as a Novel Lipid-Based Gene Nanocarrier: A Biophysical and Biochemical Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Characterization of the Intermediates and the Gemini Cationic Lipid C3(C16His)2
2.3. Preparation of Lipoplexes
2.4. Experimental Methods
3. Results
3.1. Synthesis of the Gemini Lipid (C3(C16His)2)
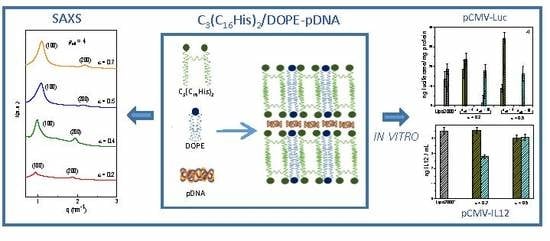
3.2. Biophysics of the C3(C16His)2/DOPE-pDNA Lipoplexes
3.3. In Vitro Biological Activity of C3(C16His)2/DOPE-pDNA Lipoplexes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| C3(C16His)2 | bis(N(τ),N(π)-bis(methyl)-histidine hexadecyl amide) propane |
| DOPE | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine |
| DPH | Diphenylhexatriene fluorescent probe |
| FBS | Fetal Bovine Serum |
| Lipo2000* | Control Lipofectamine 2000 in presence of serum FBS |
| pCMV-Luc | Plasmid DNA encoding Luciferase |
| pCMV-IL12 | Plasmid DNA encoding Interleukin-12 |
| pEGFP-C3 | Plasmid DNA encoding Green Fluorescent Protein |
| Positive effective charge of the cationic lipid | |
| Negative effective charge of plasmid DNA per base pair (bp) |
References
- Zheng, L.T.; Yi, W.J.; Su, R.C.; Liu, Q.; Zhao, Z.G. Reducible Amino Acid Based Cationic Lipids as Highly Efficient and Serum-Tolerant Gene Vectors. Chempluschem 2016, 81, 125–134. [Google Scholar] [CrossRef]
- Su, R.C.; Liu, Q.; Yi, W.J.; Zheng, L.T.; Zhao, Z.G. Lipoic acid functionalized amino acids cationic lipids as gene vectors. Bioorg. Med. Chem. 2016, 26, 4692–4697. [Google Scholar] [CrossRef] [PubMed]
- Obata, Y.; Suzuki, D.; Takeoka, S. Evaluation of cationic assemblies constructed with amino acid based lipids for plasmid DNA delivery. Bioconjugate Chem. 2008, 19, 1055–1063. [Google Scholar] [CrossRef]
- Damen, M.; Cristobal-Lecina, E.; Sanmarti, G.C.; van Dongen, S.F.M.; Garcia Rodriguez, C.L.; Dolbnya, I.P.; Nolte, R.J.M.; Feiters, M.C. Structure-delivery relationships of lysine-based gemini surfactants and their lipoplexes. Soft Matter 2014, 10, 5702–5714. [Google Scholar] [CrossRef] [PubMed]
- Midoux, P.; Pichon, C.; Yaouanc, J.J.; Jaffres, P.A. Chemical vectors for gene delivery: A current review on polymers, peptides and lipids containing histidine or imidazole as nucleic acids carriers. Br. J. Pharmacol. 2009, 157, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Dias, R.S.; Lindman, B. DNA Interaction with Polymers and Surfactants; Wiley & Sons: Hoboken, NJ, USA, 2008. [Google Scholar]
- Douat, C.; Aisenbrey, C.; Antunes, S.; Decossas, M.; Lambert, O.; Bechinger, B.; Kichler, A.; Guichard, G. A cell-penetrating foldamer with a bioreducible linkage for intracellular delivery of DNA. Angew. Chem. Int. Ed. 2015, 54, 11133–11137. [Google Scholar] [CrossRef] [PubMed]
- Kanazawa, T.; Yamazaki, M.; Fukuda, T.; Takashima, Y.; Okada, H. Versatile nuclear localization signal-based oligopeptide as a gene vector. Biol. Pharm. Bull. 2015, 38, 559–565. [Google Scholar] [CrossRef]
- Tai, Z.G.; Wang, X.Y.; Tian, J.; Gao, Y.; Zhang, L.J.; Yao, C.; Wu, X.; Zhang, W.; Zhu, Q.G.; Gao, S. Biodegradable stearylated peptide with internal disulfide bonds for efficient delivery of siRNA in vitro and in vivo. Biomacromolecules 2015, 16, 1119–1130. [Google Scholar] [CrossRef]
- Wang, X.Y.; Tai, Z.G.; Tian, J.; Zhang, W.; Yao, C.; Zhang, L.J.; Gao, Y.; Zhu, Q.G.; Gao, J.; Gao, S. Reducible chimeric polypeptide consisting of octa-D-arginine and tetra-L-histidine peptides as an efficient gene delivery vector. Int. J. Nanomed. 2015, 10, 4669–4690. [Google Scholar]
- Okuda, T.; Sugiyama, A.; Niidome, T.; Aoyagi, H. Characters of dendritic poly((L)-lysine) analogues with the terminal lysines replaced with arginines and histidines as gene carriers in vitro. Biomaterials 2004, 25, 537–544. [Google Scholar] [CrossRef]
- Bogacheva, M.; Egorova, A.; Slita, A.; Maretina, M.; Baranov, V.; Kiselev, A. Arginine-rich cross-linking peptides with different SV40 nuclear localization signal content as vectors for intranuclear DNA delivery. Bioorg. Med. Chem. Lett. 2017, 27, 4781–4785. [Google Scholar] [CrossRef] [PubMed]
- Chou, S.T.; Hom, K.; Zhang, D.; Leng, Q.; Tricoli, L.J.; Hustedt, J.M.; Lee, A.; Shapiro, M.J.; Seog, J.; Kahn, J.D.; et al. Enhanced silencing and stabilization of siRNA polyplexes by histidine-mediated hydrogen bonds. Biomaterials 2014, 35, 846–855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Z.; Zhao, D.M.; Deng, X.; Zhang, J.; Zhang, Y.M.; Yu, X.Q. Functionalized asymmetric bola-type amphiphiles for efficient gene and drug delivery. Nanomaterials 2018, 8, 115. [Google Scholar] [CrossRef] [PubMed]
- Leng, Q.X.; Chou, S.T.; Scaria, P.V.; Woodle, M.C.; Mixson, A.J. Increased tumor distribution and expression of histidine-rich plasmid polyplexes. J. Gene Med. 2014, 16, 317–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stevenson, M.; Ramos-Perez, V.; Singh, S.; Soliman, M.; Preece, J.A.; Briggs, S.S.; Read, M.L.; Seymour, L.W. Delivery of siRNA mediated by histidine-containing reducible polycations. J. Controll. Release 2008, 130, 46–56. [Google Scholar] [CrossRef]
- Kamaruzaman, K.A.; Moyle, P.M.; Toth, I. Peptide-based multicomponent oligonucleotide delivery systems: Optimisation of poly-L-lysine dendrons for plasmid DNA delivery. Int. J. Pept. Res. Ther. 2017, 23, 119–134. [Google Scholar] [CrossRef]
- Kim, J.W.; Lee, J.J.; Choi, J.S.; Kim, H.S. Electrostatically assembled dendrimer complex with a high-affinity protein binder for targeted gene delivery. Int. J. Pharm. 2018, 544, 39–45. [Google Scholar] [CrossRef]
- Wang, F.; Wang, Y.T.; Wang, H.; Shao, N.M.; Chen, Y.Y.; Cheng, Y.Y. Synergistic effect of amino acids modified on dendrimer surface in gene delivery. Biomaterials 2014, 35, 9187–9198. [Google Scholar] [CrossRef]
- Zhou, J.S.; Li, Y.; Dong, H.Q.; Yuan, H.; Ren, T.B.; Li, Y.Y. Effect of monomer sequence of poly(histidine/lysine) catiomers on gene packing capacity and delivery efficiency. RSC Adv. 2015, 5, 14138–14146. [Google Scholar] [CrossRef]
- Junquera, E.; Aicart, E. Cationic lipids as transfecting agents of DNA in gene therapy. Curr. Top. Med. Chem. 2014, 14, 649–663. [Google Scholar] [CrossRef]
- Shigeta, K.; Kawakami, S.; Higuchi, Y.; Okuda, T.; Yagi, H.; Yamashita, F.; Hashida, M. Novel histidine-conjugated galactosylated cationic liposomes for efficient hepatocyte-selective gene transfer in human hepatoma HepG2 cells. J. Controll. Release 2007, 118, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.K.; Mani, P.; Sharma, N.R.; Krishnan, A.; Kumar, V.V.; Reddy, B.S.; Chaudhuri, A.; Roy, R.P.; Sarkar, D.P. Histidylated lipid-modified Sendai viral envelopes mediate enhanced membrane fusion and potentiate targeted gene delivery. J. Biol. Chem. 2005, 280, 35399–35409. [Google Scholar] [CrossRef] [PubMed]
- Vannucci, L.; Lai, M.; Chiuppesi, F.; Ceccherini-Nelli, L.; Pistello, M. Viral vectors: A look back and ahead on gene transfer technology. New Microbiol. 2013, 36, 1–22. [Google Scholar] [PubMed]
- Mintzer, M.A.; Simanek, E.E. Nonviral vectors for gene delivery. Chem. Rev. 2008, 109, 259–302. [Google Scholar] [CrossRef] [PubMed]
- Raper, S.E.; Yudkoff, M.; Chirmule, N.; Gao, G.-P.; Nunes, F.; Haskal, Z.J.; Furth, E.E.; Propert, K.J.; Robinson, M.B.; Magosin, S. A pilot study of in vivo liver-directed gene transfer with an adenoviral vector in partial ornithine transcarbamylase deficiency. Hum. Gene Ther. 2002, 13, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Junquera, E.; Aicart, E. Recent progress in gene therapy to deliver nucleic acids with multivalent cationic vectors. Adv. Colloid Interface Sci. 2016, 233, 161–175. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Negro, M.; Kumar, K.; Barrán-Berdón, A.L.; Datta, S.; Kondaiah, P.; Junquera, E.; Bhattacharya, S.; Aicart, E. Efficient cellular knockdown mediated by siRNA nanovectors of gemini cationic lipids having delocalizable headgroups and oligo-oxyethylene spacers. ACS Appl. Mater. Interfaces 2016, 8, 22113–22126. [Google Scholar] [CrossRef]
- Barrán-Berdón, A.L.; Misra, S.K.; Datta, S.; Muñoz-Úbeda, M.; Kondaiah, P.; Junquera, E.; Bhattacharya, S.; Aicart, E. Cationic gemini lipids containing polyoxyethylene spacers as improved transfecting agents of plasmid DNA in cancer cells. J. Mater. Chem. B 2014, 2, 4640–4652. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Bajaj, A. Advances in gene delivery through molecular design of cationic lipids. Chem. Commun. 2009, 31, 4632–4656. [Google Scholar] [CrossRef]
- Kirby, A.J.; Camilleri, P.; Engberts, J.; Feiters, M.C.; Nolte, R.J.M.; Soderman, O.; Bergsma, M.; Bell, P.C.; Fielden, M.L.; Rodriguez, C.L.G.; et al. Gemini surfactants: New synthetic vectors for gene transfection. Angew. Chem. Int. Ed. 2003, 42, 1448–1457. [Google Scholar] [CrossRef]
- Muñoz-Úbeda, M.; Misra, S.K.; Barrán-Berdón, A.L.; Datta, S.; Aicart-Ramos, C.; Castro-Hartmann, P.; Kondaiah, P.; Junquera, E.; Bhattacharya, S.; Aicart, E. How does the spacer length of cationic gemini lipids influence the lipoplex formation with plasmid DNA? Physicochemical and biochemical characterizations and their relevance in gene therapy. Biomacromolecules 2012, 13, 3926–3937. [Google Scholar] [CrossRef] [PubMed]
- Misra, S.K.; Muñoz-Úbeda, M.; Datta, S.; Barrán-Berdón, A.L.; Aicart-Ramos, C.; Castro-Hartmann, P.; Kondaiah, P.; Junquera, E.; Bhattacharya, S.; Aicart, E. Effects of a delocalizable cation on the headgroup of gemini lipids on the lipoplex-type nano-aggregates directly formed from plasmid DNA. Biomacromolecules 2013, 14, 3951–3963. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Xu, X.; Li, H.; Jian, Y.; Wang, G.; He, B.; Gu, Z. Components simulation of viral envelope via amino acid modified chitosans for efficient nucleic acid delivery: In vitro and in vivo study. Adv. Funct. Mater. 2013, 23, 2691–2699. [Google Scholar] [CrossRef]
- Kono, K.; Ikeda, R.; Tsukamoto, K.; Yuba, E.; Kojima, C.; Harada, A. Polyamidoamine dendron-bearing lipids as a nonviral vector: Influence of dendron generation. Bioconjugate Chem. 2012, 23, 871–879. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, J.; Xiao, C.; Li, M.; Tian, H.; Chen, X. Cationic dendron-bearing lipids: Investigating structure–activity relationships for small interfering RNA delivery. Biomacromolecules 2013, 14, 4289–4300. [Google Scholar] [CrossRef] [PubMed]
- Karmali, P.P.; Majeti, B.K.; Sreedhar, B.; Chaudhuri, A. In vitro gene transfer efficacies and serum compatibility profiles of novel mono-, di-, and tri-histidinylated cationic transfection lipids: A structure-activity investigation. Bioconjugate Chem. 2006, 17, 159–171. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.V.; Pichon, C.; Refregiers, M.; Guerin, B.; Midoux, P.; Chaudhuri, A. Single histidine residue in head-group region is sufficient to impart remarkable gene transfection properties to cationic lipids: Evidence for histidine-mediated membrane fusion at acidic pH. Gene Ther. 2003, 10, 1206–1215. [Google Scholar] [CrossRef]
- Hwang, H.S.; Hu, J.; Na, K.; Bae, Y.H. Role of polymeric endosomolytic agents in gene transfection: A comparative study of poly(L-lysine) grafted with monomeric L-histidine analogue and poly(L-histidine). Biomacromolecules 2014, 15, 3577–3586. [Google Scholar] [CrossRef]
- Aldawsari, H.; Edrada-Ebel, R.; Blatchford, D.R.; Tate, R.J.; Tetley, L.; Dufès, C. Enhanced gene expression in tumors after intravenous administration of arginine-, lysine-and leucine-bearing polypropylenimine polyplex. Biomaterials 2011, 32, 5889–5899. [Google Scholar] [CrossRef]
- Barrán-Berdón, A.L.; Muñoz-Úbeda, M.; Aicart-Ramos, C.; Pérez, L.; Infante, M.R.; Castro-Hartmann, P.; Martín-Molina, A.; Aicart, E.; Junquera, E. Ribbon-type and cluster-type lipoplexes constituted by a chiral lysine based cationic gemini lipid and plasmid DNA. Soft Matter 2012, 8, 7368–7380. [Google Scholar] [CrossRef]
- Kim, T.I.; Baek, J.U.; Bai, C.Z.; Park, J.S. Arginine-conjugated polypropylenimine dendrimer as a non-toxic and efficient gene delivery carrier. Biomaterials 2007, 28, 2061–2067. [Google Scholar] [CrossRef] [PubMed]
- Vigneron, J.P.; Oudrhiri, N.; Fauquet, M.; Vergely, L.; Bradley, J.C.; Basseville, M.; Lehn, P.; Lehn, J.M. Guanidinium-cholesterol cationic lipids: Efficient vectors for the transfection of eukaryotic cells. Proc. Nat. Acad. Sci. USA 1996, 93, 9682–9686. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Nie, Y.; Zhu, R.; Shi, S.; Luo, K.; He, B.; Yang, Y.; Yang, J.; Gu, Z. Preparation and gene delivery of alkaline amino acids-based cationic liposomes. Arch. Pharm. Res. 2008, 31, 924–931. [Google Scholar] [CrossRef] [PubMed]
- Kichler, A.; Leborgne, C.; März, J.; Danos, O.; Bechinger, B. Histidine-rich amphipathic peptide antibiotics promote efficient delivery of DNA into mammalian cells. Proc. Nat. Acad. Sci. USA 2003, 100, 1564–1568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heyes, J.A.; Niculescu-Duvaz, D.; Cooper, R.G.; Springer, C.J. Synthesis of novel cationic lipids: Effect of structural modification on the efficiency of gene transfer. J. Med. Chem. 2002, 45, 99–114. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Yue, D.; Nie, Y.; Xu, X.H.; He, Y.Y.; Zhang, S.Y.; Wagner, E.; Gu, Z.W. Specially-Made Lipid-Based Assemblies for Improving Transmembrane Gene Delivery: Comparison of Basic Amino Acid Residue Rich Periphery. Mol. Pharm. 2016, 13, 1809–1821. [Google Scholar] [CrossRef] [PubMed]
- Ju, J.; Huan, M.L.; Wan, N.; Qiu, H.; Zhou, S.Y.; Zhang, B.L. Novel cholesterol-based cationic lipids as transfecting agents of DNA for efficient gene delivery. Int. J. Mol. Sci. 2015, 16, 5666–5681. [Google Scholar] [CrossRef]
- Dobbs, W.; Heinrich, B.; Bourgogne, C.; Donnio, B.; Terazzi, E.; Bonnet, M.E.; Stock, F.; Erbacher, P.; Bolcato Bellemin, A.L.; Douce, L. Mesomorphic imidazolium salts: New vectors for efficient siRNA transfection. J. Am. Chem. Soc. 2009, 131, 13338–13346. [Google Scholar] [CrossRef]
- Zhou, T.; Llizo, A.; Li, P.; Wang, C.; Guo, Y.; Ao, M.; Bai, L.; Wang, C.; Yang, Y.; Xu, G. High transfection efficiency of homogeneous DNA nanoparticles induced by imidazolium gemini surfactant as nonviral vector. J. Phys. Chem. C 2013, 117, 26573–26581. [Google Scholar] [CrossRef]
- Kumar, K.; Barrán-Berdón, A.L.; Datta, S.; Muñoz-Úbeda, M.; Aicart-Ramos, C.; Kondaiah, P.; Junquera, E.; Bhattacharya, S.; Aicart, E. A delocalizable cationic headgroup together with an oligo-oxyethylene spacer in gemini cationic lipids improves their biological activity as vectors of plasmid DNA. J. Mater. Chem. B 2015, 3, 1495–1506. [Google Scholar] [CrossRef]
- Martinez-Negro, M.; Guerrero-Martinez, A.; Garcia-Rio, L.; Domenech, O.; Aicart, E.; de Ilarduya, C.T.; Junquera, E. Multidisciplinary approach to the transfection of plasmid DNA by a nonviral nanocarrier based on a gemini-bolaamphiphilic hybrid lipid. ACS Omega 2018, 3, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Pulido, A.; Aicart, E.; Junquera, E. Electrochemical and spectroscopic study of octadecyltrimethylammonium bromide/DNA surfoplexes. Langmuir 2009, 25, 4402–4411. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, T.; Kamel, A.O.; Wettig, S.D. Interactions between DNA and Gemini surfactant: Impact on gene therapy: Part I. Nanomedicine 2016, 11, 289–306. [Google Scholar] [CrossRef] [PubMed]
- Pinazo, A.; Manresa, M.; Marques, A.; Bustelo, M.; Espuny, M.; Perez, L. Amino acid–based surfactants: New antimicrobial agents. Adv. Colloid Interface Sci. 2016, 228, 17–39. [Google Scholar] [CrossRef] [PubMed]
- Pinazo, A.; Angelet, M.; Pons, R.; Lozano, M.; Infante, M.R.; Perez, L. Lysine-bisglycidol conjugates as novel lysine cationic surfactants. Langmuir 2009, 25, 7803–7814. [Google Scholar] [CrossRef] [PubMed]
- Colomer, A.; Pinazo, A.; García, M.T.; Mitjans, M.; Viardell, M.P.; Infante, M.R.; Martínez, V.N.; Pérez, L. pH-sensitive surfactants from lysine: Assessment of their cytotoxicity and environmental behavior. Langmuir 2012, 28, 5900–5912. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, A.M.; Morais, C.M.; Silva, S.G.; Marques, E.F.; de Lima, M.C.P.; Jurado, M.A.S. Bis-quaternary gemini surfactants as components of nonviral gene delivery systems: A comprehensive study from physicochemical properties to membrane interactions. Int. J. Pharm. 2014, 474, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Satyal, U.; Draghici, B.; Dragic, L.L.; Zhang, Q.; Norris, K.W.; Madesh, M.; Brailoiu, E.; Ilies, M.A. Interfacially engineered pyridinium pseudogemini surfactants as versatile and efficient supramolecular delivery systems for DNA, siRNA, and mRNA. ACS Appl. Mater. Interfaces 2017, 9, 29481–29495. [Google Scholar] [CrossRef]
- Bustelo, M.; Pinazo, A.; Manresa, M.; Mitjans, M.; Vinardell, M.; Perez, L. Monocatenary histidine-based surfactants: Role of the alkyl chain length in antimicrobial activity and their selectivity over red blood cells. Colloids Surf. A 2017, 532, 501–509. [Google Scholar] [CrossRef]
- Muñoz-Úbeda, M.; Misra, S.K.; Barrán-Berdón, A.L.; Aicart-Ramos, C.; Sierra, M.B.; Biswas, J.; Kondaiah, P.; Junquera, E.; Bhattacharya, S.; Aicart, E. Why is less cationic lipid required to prepare lipoplexes from plasmid DNA than linear DNA in gene therapy? J. Am. Chem. Soc. 2011, 133, 18014–18017. [Google Scholar] [CrossRef]
- Foldvari, M.; Badea, I.; Wettig, S.; Verrall, R.; Bagonluri, M. Structural characterization of novel gemini non-viral DNA delivery systems for cutaneous gene therapy. J. Exp. Nanosci. 2006, 1, 165–176. [Google Scholar] [CrossRef]
- Lyubchenko, Y.L.; Shlyakhtenko, L.S. Visualization of supercoiled DNA with atomic force microscopy in situ. Proc. Natl. Acad. Sci. USA 1997, 94, 496–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barran-Berdon, A.L.; Martinez-Negro, M.; Garcia-Rio, L.; Domenech, O.; de Ilarduya, C.T.; Aicart, E.; Junquera, E. A biophysical study of gene nanocarriers formed by anionic/zwitterionic mixed lipids and pillar-5-arene polycationic macrocycles. J. Mater. Chem. B 2017, 5, 3122–3131. [Google Scholar] [CrossRef]
- Tanford, C. The Hydrophobic Effect: Formation of Micelles and Biological Membranes; Wiley & Sons: New York, NY, USA, 1980. [Google Scholar]
- Tanford, C. Micelle shape and size. J. Phys. Chem. 1972, 76, 3020–3024. [Google Scholar] [CrossRef]
- Tanford, C. Theory of micelle formation in aqueous solutions. J. Phys. Chem. 1974, 78, 2469–2479. [Google Scholar] [CrossRef]
- Borenstain, V.; Barenholz, Y. Characterization of liposomes and other lipid assemblies by multiprobe fluorescence polarization. Chem. Phys. Lipids 1993, 64, 117–127. [Google Scholar] [CrossRef]
- Phillips, J.N. The energetics of micelle formation. Trans. Faraday Soc. 1955, 51, 561–569. [Google Scholar] [CrossRef]









| Lipoplex | ρeff = 4 | ρeff = 10 | ||
|---|---|---|---|---|
| Dh (nm) | PDI | Dh (nm) | PDI | |
| α = 0.2 | ||||
| C3(C16His)2/DOPE-pEGFP-C3 | 170 | 0.17 | 136 | 0.11 |
| C3(C16His)2/DOPE-pCMV-Luc | 177 | 0.27 | 119 | 0.15 |
| α = 0.5 | ||||
| C3(C16His)2/DOPE-pEGFP-C3 | 147 | 0.12 | 177 | 0.40 |
| C3(C16His)2/DOPE-pCMV-Luc | 154 | 0.19 | 185 | 0.26 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Negro, M.; Blanco-Fernández, L.; Tentori, P.M.; Pérez, L.; Pinazo, A.; Tros de Ilarduya, C.; Aicart, E.; Junquera, E. A Gemini Cationic Lipid with Histidine Residues as a Novel Lipid-Based Gene Nanocarrier: A Biophysical and Biochemical Study. Nanomaterials 2018, 8, 1061. https://doi.org/10.3390/nano8121061
Martínez-Negro M, Blanco-Fernández L, Tentori PM, Pérez L, Pinazo A, Tros de Ilarduya C, Aicart E, Junquera E. A Gemini Cationic Lipid with Histidine Residues as a Novel Lipid-Based Gene Nanocarrier: A Biophysical and Biochemical Study. Nanomaterials. 2018; 8(12):1061. https://doi.org/10.3390/nano8121061
Chicago/Turabian StyleMartínez-Negro, María, Laura Blanco-Fernández, Paolo M. Tentori, Lourdes Pérez, Aurora Pinazo, Conchita Tros de Ilarduya, Emilio Aicart, and Elena Junquera. 2018. "A Gemini Cationic Lipid with Histidine Residues as a Novel Lipid-Based Gene Nanocarrier: A Biophysical and Biochemical Study" Nanomaterials 8, no. 12: 1061. https://doi.org/10.3390/nano8121061