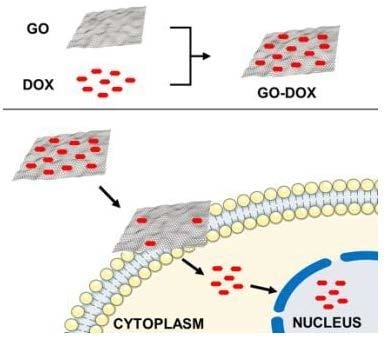
Mechanistic Insights into the Release of Doxorubicin from Graphene Oxide in Cancer Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of GO-DOX Complexes
2.2. UV-VIS Experiments
2.3. Dynamic Light Scattering
2.4. Cell Culture
2.5. Cell Metabolic Activity Experiments
2.6. Confocal Imaging and Phasor-FLIM Experiments
2.7. Statistical Analysis
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Singal, P.K.; Iliskovic, N. Doxorubicin-induced cardiomyopathy. N. Engl. J. Med. 1998, 339, 900–905. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.-L.; Li, Q.-X.; Xia, L.; Li, J.; Shao, L. Differential effects of dihydropyridine calcium antagonists on doxorubicin-induced nephrotoxicity in rats. Toxicology 2007, 231, 81–90. [Google Scholar] [CrossRef]
- Barenholz, Y.; Cohen, R. Rational design of amphiphile-based drug carriers and sterically stabilized carriers. J. Liposome Res. 1995, 5, 905–932. [Google Scholar] [CrossRef]
- Barenholz, Y.C. Doxil®—The first fda-approved nano-drug: Lessons learned. J. Control. Release 2012, 160, 117–134. [Google Scholar] [CrossRef]
- Caracciolo, G. Clinically approved liposomal nanomedicines: Lessons learned from the biomolecular corona. Nanoscale 2018, 10, 4167–4172. [Google Scholar] [CrossRef] [PubMed]
- Caracciolo, G.; Palchetti, S.; Digiacomo, L.; Chiozzi, R.Z.; Capriotti, A.L.; Amenitsch, H.; Tentori, P.M.; Palmieri, V.; Papi, M.; Cardarelli, F. Human biomolecular corona of liposomal doxorubicin: The overlooked factor in anticancer drug delivery. ACS Appl. Mater. Interfaces 2018, 10, 22951–22962. [Google Scholar] [CrossRef]
- Seynhaeve, A.L.; Dicheva, B.M.; Hoving, S.; Koning, G.A.; ten Hagen, T.L. Intact Doxil is taken up intracellularly and released doxorubicin sequesters in the lysosome: Evaluated by in vitro/in vivo live cell imaging. J. Control. Release 2013, 172, 330–340. [Google Scholar] [CrossRef]
- Chung, C.; Kim, Y.-K.; Shin, D.; Ryoo, S.-R.; Hong, B.H.; Min, D.-H. Biomedical applications of graphene and graphene oxide. Acc. Chem. Res. 2013, 46, 2211–2224. [Google Scholar] [CrossRef]
- Liu, J.; Cui, L.; Losic, D. Graphene and graphene oxide as new nanocarriers for drug delivery applications. Acta Biomater. 2013, 9, 9243–9257. [Google Scholar] [CrossRef]
- Liu, Z.; Robinson, J.T.; Sun, X.; Dai, H. PEGylated nanographene oxide for delivery of water-insoluble cancer drugs. J. Am. Chem. Soc. 2008, 130, 10876–10877. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Liu, Z.; Welsher, K.; Robinson, J.T.; Goodwin, A.; Zaric, S.; Dai, H. Nano-graphene oxide for cellular imaging and drug delivery. Nano Res. 2008, 1, 203–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, S.; Zhao, X.; Li, Y.; Du, Q.; Sun, J.; Wang, Y.; Wang, X.; Xia, Y.; Wang, Z.; Xia, L. Adsorption properties of doxorubicin hydrochloride onto graphene oxide: Equilibrium, kinetic and thermodynamic studies. Materials 2013, 6, 2026–2042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, S.-Y.; An, S.S.A.; Hulme, J. Current applications of graphene oxide in nanomedicine. Int. J. Nanomed. 2015, 10, 9. [Google Scholar]
- Shareena, T.P.D.; McShan, D.; Dasmahapatra, A.K.; Tchounwou, P.B. A review on graphene-based nanomaterials in biomedical applications and risks in environment and health. Nano-Micro Lett. 2018, 10, 53. [Google Scholar] [CrossRef] [PubMed]
- Lankelma, J.; Dekker, H.; Luque, R.F.; Luykx, S.; Hoekman, K.; Van Der Valk, P.; Van Diest, P.J.; Pinedo, H.M. Doxorubicin gradients in human breast cancer. Clin. Cancer Res. 1999, 5, 1703–1707. [Google Scholar] [PubMed]
- Zhao, X.; Liu, L.; Li, X.; Zeng, J.; Jia, X.; Liu, P. Biocompatible graphene oxide nanoparticle-based drug delivery platform for tumor microenvironment-responsive triggered release of doxorubicin. Langmuir 2014, 30, 10419–10429. [Google Scholar] [CrossRef]
- Khan, M.S.; Hwang, J.; Lee, K.; Choi, Y.; Seo, Y.; Jeon, H.; Hong, J.W.; Choi, J. Anti-tumor drug-loaded oxygen nanobubbles for the degradation of HIF-1α and the upregulation of reactive oxygen species in tumor cells. Cancers 2019, 11, 1464. [Google Scholar] [CrossRef] [Green Version]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Gies, V.; Lopinski, G.; Augustine, J.; Cheung, T.; Kodra, O.; Zou, S. The impact of processing on the cytotoxicity of graphene oxide. Nanoscale Adv. 2019, 1, 817–826. [Google Scholar] [CrossRef] [Green Version]
- Hashemi, M.; Yadegari, A.; Yazdanpanah, G.; Omidi, M.; Jabbehdari, S.; Haghiralsadat, F.; Yazdian, F.; Tayebi, L. Normalization of doxorubicin release from graphene oxide: New approach for optimization of effective parameters on drug loading. Biotechnol. Appl. Biochem. 2017, 64, 433–442. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, X.; Liu, Z.; Ma, Y.; Huang, Y.; Chen, Y. High-efficiency loading and controlled release of doxorubicin hydrochloride on graphene oxide. J. Phys. Chem. C 2008, 112, 17554–17558. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, W.; Kong, T.; Su, R.; Li, N.; Song, Q.; Tang, M.; Liu, L.; Cheng, G. Tailoring the interlayer interaction between doxorubicin-loaded graphene oxide nanosheets by controlling the drug content. Carbon 2013, 51, 164–172. [Google Scholar] [CrossRef]
- Choudhary, P.; Parandhaman, T.; Ramalingam, B.; Duraipandy, N.; Kiran, M.S.; Das, S.K. Fabrication of nontoxic reduced graphene oxide protein nanoframework as sustained antimicrobial coating for biomedical application. ACS Appl. Mater. Interfaces 2017, 9, 38255–38269. [Google Scholar] [CrossRef] [PubMed]
- Javanbakht, S.; Namazi, H. Doxorubicin loaded carboxymethyl cellulose/graphene quantum dot nanocomposite hydrogel films as a potential anticancer drug delivery system. Mater. Sci. Eng. C 2018, 87, 50–59. [Google Scholar] [CrossRef]
- Digman, M.A.; Caiolfa, V.R.; Zamai, M.; Gratton, E. The phasor approach to fluorescence lifetime imaging analysis. Biophys. J. 2008, 94, L14–L16. [Google Scholar] [CrossRef] [Green Version]
- Shah, S.; Chandra, A.; Kaur, A.; Sabnis, N.; Lacko, A.; Gryczynski, Z.; Fudala, R.; Gryczynski, I. Fluorescence properties of doxorubicin in PBS buffer and PVA films. J. Photochem. Photobiol. B Biol. 2017, 170, 65–69. [Google Scholar] [CrossRef] [Green Version]
- Vovusha, H.; Banerjee, D.; Yadav, M.K.; Perrozzi, F.; Ottaviano, L.; Sanyal, S.; Sanyal, B. Binding characteristics of anticancer drug doxorubicin with two-dimensional graphene and graphene oxide: Insights from density functional theory calculations and fluorescence spectroscopy. J. Phys. Chem. C 2018, 122, 21031–21038. [Google Scholar] [CrossRef]
- Stringari, C.; Cinquin, A.; Cinquin, O.; Digman, M.A.; Donovan, P.J.; Gratton, E. Phasor approach to fluorescence lifetime microscopy distinguishes different metabolic states of germ cells in a live tissue. Proc. Natl. Acad. Sci. USA 2011, 108, 13582–13587. [Google Scholar] [CrossRef] [Green Version]
- Basuki, J.S.; Duong, H.T.; Macmillan, A.; Erlich, R.B.; Esser, L.; Akerfeldt, M.C.; Whan, R.M.; Kavallaris, M.; Boyer, C.; Davis, T.P. Using fluorescence lifetime imaging microscopy to monitor theranostic nanoparticle uptake and intracellular doxorubicin release. ACS Nano 2013, 7, 10175–10189. [Google Scholar] [CrossRef]
- Caccamo, D.; Currò, M.; Ientile, R.; Verderio, E.A.; Scala, A.; Mazzaglia, A.; Pennisi, R.; Musarra-Pizzo, M.; Zagami, R.; Neri, G. Intracellular fate and impact on gene expression of doxorubicin/cyclodextrin-graphene nanomaterials at sub-toxic concentration. Int. J. Mol. Sci. 2020, 21, 4891. [Google Scholar] [CrossRef]
- Zhu, J.; Xu, M.; Gao, M.; Zhang, Z.; Xu, Y.; Xia, T.; Liu, S. Graphene oxide induced perturbation to plasma membrane and cytoskeletal meshwork sensitize cancer cells to chemotherapeutic agents. ACS Nano 2017, 11, 2637–2651. [Google Scholar] [CrossRef]
- Zhang, X.; Shastry, S.; Bradforth, S.E.; Nadeau, J.L. Nuclear uptake of ultrasmall gold-doxorubicin conjugates imaged by fluorescence lifetime imaging microscopy (FLIM) and electron microscopy. Nanoscale 2015, 7, 240–251. [Google Scholar] [CrossRef] [PubMed]
- Suarasan, S.; Craciun, A.-M.; Licarete, E.; Focsan, M.; Magyari, K.; Astilean, S. Intracellular dynamic disentangling of doxorubicin release from luminescent nanogold carriers by fluorescence lifetime imaging microscopy (FLIM) under two-photon excitation. ACS Appl. Mater. Interfaces 2019, 11, 7812–7822. [Google Scholar] [CrossRef] [PubMed]
- Barua, S.; Mitragotri, S. Challenges associated with penetration of nanoparticles across cell and tissue barriers: A review of current status and future prospects. Nano Today 2014, 9, 223–243. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yu, D.; Dai, R.; Fu, D.; Li, W.; Guo, Z.; Cui, C.; Xu, J.; Shen, S.; Ma, K. PEGylated doxorubicin cloaked nano-graphene oxide for dual-responsive photochemical therapy. Int. J. Pharm. 2019, 557, 66–73. [Google Scholar] [CrossRef]
- Huber, L.A.; Pereira, T.A.; Ramos, D.N.; Rezende, L.C.; Emery, F.S.; Sobral, L.M.; Leopoldino, A.M.; Lopez, R.F. Topical skin cancer therapy using doxorubicin-loaded cationic lipid nanoparticles and iontophoresis. J. Biomed. Nanotechnol. 2015, 11, 1975–1988. [Google Scholar] [CrossRef]
- Zhao, N.; Woodle, M.C.; Mixson, A.J. Advances in delivery systems for doxorubicin. J. Nanomed. Nanotechnol. 2018, 9, 519. [Google Scholar] [CrossRef]
- Capriotti, A.L.; Caracciolo, G.; Caruso, G.; Cavaliere, C.; Pozzi, D.; Samperi, R.; Laganà, A. Analysis of plasma protein adsorption onto DC-Chol-DOPE cationic liposomes by HPLC-CHIP coupled to a Q-TOF mass spectrometer. Anal. Bioanal. Chem. 2010, 398, 2895–2903. [Google Scholar] [CrossRef]
- Palchetti, S.; Pozzi, D.; Capriotti, A.L.; La Barbera, G.; Chiozzi, R.Z.; Digiacomo, L.; Peruzzi, G.; Caracciolo, G.; Laganà, A. Influence of dynamic flow environment on nanoparticle-protein corona: From protein patterns to uptake in cancer cells. Colloids Surf. B Biointerfaces 2017, 153, 263–271. [Google Scholar] [CrossRef]
- Caracciolo, G.; Caputo, D.; Pozzi, D.; Colapicchioni, V.; Coppola, R. Size and charge of nanoparticles following incubation with human plasma of healthy and pancreatic cancer patients. Colloids Surf. B Biointerfaces 2014, 123, 673–678. [Google Scholar] [CrossRef] [PubMed]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quagliarini, E.; Di Santo, R.; Pozzi, D.; Tentori, P.; Cardarelli, F.; Caracciolo, G. Mechanistic Insights into the Release of Doxorubicin from Graphene Oxide in Cancer Cells. Nanomaterials 2020, 10, 1482. https://doi.org/10.3390/nano10081482
Quagliarini E, Di Santo R, Pozzi D, Tentori P, Cardarelli F, Caracciolo G. Mechanistic Insights into the Release of Doxorubicin from Graphene Oxide in Cancer Cells. Nanomaterials. 2020; 10(8):1482. https://doi.org/10.3390/nano10081482
Chicago/Turabian StyleQuagliarini, Erica, Riccardo Di Santo, Daniela Pozzi, Paolo Tentori, Francesco Cardarelli, and Giulio Caracciolo. 2020. "Mechanistic Insights into the Release of Doxorubicin from Graphene Oxide in Cancer Cells" Nanomaterials 10, no. 8: 1482. https://doi.org/10.3390/nano10081482