Unrevealing the Potential of Sansevieria trifasciata Prain Fraction for the Treatment of Androgenetic Alopecia by Inhibiting Androgen Receptors Based on LC-MS/MS Analysis, and In-Silico Studies
Abstract
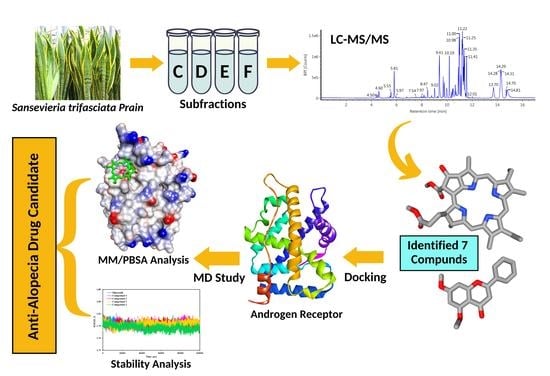
:1. Introduction
2. Results
2.1. Phytochemical Constituent by LC-MS/MS Analysis
2.2. Molecular Docking Simulation
2.3. Molecular Dynamics Simulation
2.4. Binding Energy Calculation of the Complexes
2.5. Pharmacokinetic and Toxicity Prediction
3. Discussion
4. Materials and Methods
4.1. Extraction and Separation Compounds
4.2. LC-MS/MS Analysis
4.3. Molecular Docking Simulation
4.4. Molecular Dynamics Simulation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Billero, V.; Miteva, M. Traction Alopecia: The Root of the Problem. Clin. Cosmet. Investig. Dermatol. 2018, 11, 149–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azzouni, F.; Godoy, A.; Li, Y.; Mohler, J. The 5 Alpha-Reductase Isozyme Family: A Review of Basic Biology and Their Role in Human Diseases. Adv. Urol. 2012, 2012, 530121. [Google Scholar] [CrossRef] [Green Version]
- Otberg, N.; Finner, A.M.; Shapiro, J. Androgenetic Alopecia. Endocrinol. Metab. Clin. N. Am. 2007, 36, 379–398. [Google Scholar] [CrossRef] [PubMed]
- Trüeb, R.M. Androgenetic Alopecia BT—European Handbook of Dermatological Treatments; Katsambas, A.D., Lotti, T.M., Dessinioti, C., D’Erme, A.M., Eds.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 55–65. ISBN 978-3-662-45139-7. [Google Scholar]
- Norwood, O.T. Incidence of Female Androgenetic Alopecia (Female Pattern Alopecia). Dermatol. Surg. 2001, 27, 53–54. [Google Scholar] [PubMed]
- Kaliyadan, F.; Nambiar, A.; Vijayaraghavan, S. Androgenetic Alopecia: An Update. Indian J. Dermatol. Venereol. Leprol. 2013, 79, 613–625. [Google Scholar] [CrossRef]
- Dhurat, R.; Sharma, A.; Rudnicka, L.; Kroumpouzos, G.; Kassir, M.; Galadari, H.; Wollina, U.; Lotti, T.; Golubovic, M.; Binic, I.; et al. 5-Alpha Reductase Inhibitors in Androgenetic Alopecia: Shifting Paradigms, Current Concepts, Comparative Efficacy, and Safety. Dermatol. Ther. 2020, 33, e13379. [Google Scholar] [CrossRef] [PubMed]
- Inui, S.; Itami, S. Androgen Actions on the Human Hair Follicle: Perspectives. Exp. Dermatol. 2013, 22, 168–171. [Google Scholar] [CrossRef] [PubMed]
- Madersbacher, S.; Sampson, N.; Culig, Z. Pathophysiology of Benign Prostatic Hyperplasia and Benign Prostatic Enlargement: A Mini-Review. Gerontology 2019, 65, 458–464. [Google Scholar] [CrossRef]
- Jain, R.; De-Eknamkul, W. Potential Targets in the Discovery of New Hair Growth Promoters for Androgenic Alopecia. Expert Opin. Ther. Targets 2014, 18, 787–806. [Google Scholar] [CrossRef]
- Yun, S.-I.; Lee, S.-K.; Goh, E.-A.; Kwon, O.S.; Choi, W.; Kim, J.; Lee, M.S.; Choi, S.J.; Lim, S.S.; Moon, T.K.; et al. Weekly Treatment with SAMiRNA Targeting the Androgen Receptor Ameliorates Androgenetic Alopecia. Sci. Rep. 2022, 12, 1607. [Google Scholar] [CrossRef]
- Chanprapaph, K.; Sutharaphan, T.; Suchonwanit, P. Scalp Biophysical Characteristics in Males with Androgenetic Alopecia: A Comparative Study with Healthy Controls. Clin. Interv. Aging 2021, 16, 781–787. [Google Scholar] [CrossRef]
- Davey, R.A.; Grossmann, M. Androgen Receptor Structure, Function and Biology: From Bench to Bedside. Clin. Biochem. Rev. 2016, 37, 3–15. [Google Scholar]
- Inui, S.; Itami, S. Molecular Basis of Androgenetic Alopecia: From Androgen to Paracrine Mediators through Dermal Papilla. J. Dermatol. Sci. 2011, 61, 1–6. [Google Scholar] [CrossRef]
- Hamada, K.; Thornton, M.J.; Laing, I.; Messenger, A.G.; Randall, V.A. The Metabolism of Testosterone by Dermal Papilla Cells Cultured from Human Pubic and Axillary Hair Follicles Concurs with Hair Growth in 5α-Reductase Deficiency. J. Investig. Dermatol. 1996, 106, 1017–1022. [Google Scholar] [CrossRef] [Green Version]
- Santos, Z.; Avci, P.; Hamblin, M.R. Drug Discovery for Alopecia: Gone Today, Hair Tomorrow. Expert Opin. Drug Discov. 2015, 10, 269–292. [Google Scholar] [CrossRef] [Green Version]
- Kelly, Y.; Blanco, A.; Tosti, A. Androgenetic Alopecia: An Update of Treatment Options. Drugs 2016, 76, 1349–1364. [Google Scholar] [CrossRef]
- Ahmad, R.G.E.-M. Pharmacological Treatment of Alopecia; IntechOpen: Rijeka, Croatia, 2018; Chapter 4; ISBN 978-1-78984-344-6. [Google Scholar]
- Wang, Q.; Carroll, J.S.; Brown, M. Spatial and Temporal Recruitment of Androgen Receptor and Its Coactivators Involves Chromosomal Looping and Polymerase Tracking. Mol. Cell 2005, 19, 631–642. [Google Scholar] [CrossRef]
- Messenger, A.G.; Rundegren, J. Minoxidil: Mechanisms of Action on Hair Growth. Br. J. Dermatol. 2004, 150, 186–194. [Google Scholar] [CrossRef]
- Sonthalia, S. Hair Restoration in Androgenetic Alopecia: Looking Beyond Minoxidil, Finasteride and Hair Transplantation. J. Cosmetol Trichol. 2016, 2, 1–13. [Google Scholar] [CrossRef]
- Hsu, C.-L.; Liu, J.-S.; Lin, A.-C.; Yang, C.-H.; Chung, W.-H.; Wu, W.-G. Minoxidil May Suppress Androgen Receptor-Related Functions. Oncotarget 2014, 5, 2187–2197. [Google Scholar] [CrossRef] [Green Version]
- Van Royen, M.E.; van Cappellen, W.A.; de Vos, C.; Houtsmuller, A.B.; Trapman, J. Stepwise Androgen Receptor Dimerization. J. Cell Sci. 2012, 125, 1970–1979. [Google Scholar] [CrossRef] [Green Version]
- Mella, J.M.; Perret, M.C.; Manzotti, M.; Catalano, H.N.; Guyatt, G. Efficacy and Safety of Finasteride Therapy for Androgenetic Alopecia: A Systematic Review. Arch. Dermatol. 2010, 146, 1141–1150. [Google Scholar] [CrossRef]
- Avram, M.R.; Leonard, R.T.; Epstein, E.S.; Williams, J.L.; Bauman, A.J. The Current Role of Laser/Light Sources in the Treatment of Male and Female Pattern Hair Loss. J. Cosmet. Laser Ther. 2007, 9, 27–28. [Google Scholar] [CrossRef]
- Van Zuuren, E.J.; Fedorowicz, Z.; Schoones, J. Interventions for Female Pattern Hair Loss. Cochrane Database Syst. Rev. 2016, 5, 1–189. [Google Scholar] [CrossRef]
- Suchonwanit, P.; Thammarucha, S.; Leerunyakul, K. Minoxidil and Its Use in Hair Disorders: A Review. Drug Des. Devel. Ther. 2019, 13, 2777–2786. [Google Scholar] [CrossRef] [Green Version]
- Thornton, M.J.; Taylor, A.H.; Mulligan, K.; Al-Azzawi, F.; Lyon, C.C.; O’Driscoll, J.; Messenger, A.G. The Distribution of Estrogen Receptor β Is Distinct to That of Estrogen Receptor α and the Androgen Receptor in Human Skin and the Pilosebaceous Unit. J. Investig. Dermatol. Symp. Proc. 2003, 8, 100–103. [Google Scholar] [CrossRef] [Green Version]
- Hosking, A.-M.; Juhasz, M.; Atanaskova Mesinkovska, N. Complementary and Alternative Treatments for Alopecia: A Comprehensive Review. Ski. Appendage Disord. 2019, 5, 72–89. [Google Scholar] [CrossRef]
- Mathur, S.; Hoskins, C. Drug Development: Lessons from Nature. Biomed. Rep. 2017, 6, 612–614. [Google Scholar] [CrossRef] [Green Version]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Orhan, I.E.; Banach, M.; Rollinger, J.M.; Barreca, D.; Weckwerth, W.; Bauer, R.; Bayer, E.A.; et al. Natural Products in Drug Discovery: Advances and Opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef]
- Rahmatullah, M.; Khatun, Z.; Hasan, A.; Parvin, W.; Moniruzzaman, M.; Khatun, A.; Mahal, M.J.; Bhuiyan, M.S.A.; Mou, S.M.; Jahan, R. Survey and Scientific Evaluation of Medicinal Plants Used by the Pahan and Teli Tribal Communities of Natore District, Bangladesh. Afr. J. Tradit. Complement. Altern. Med. 2012, 9, 366–373. [Google Scholar] [CrossRef] [Green Version]
- Dewatisari, W.F.; Rumiyanti, L.; Rakhmawati, I. Rendemen dan Skrining Fitokimia pada Ekstrak Daun Sanseviera sp. Rendemen and Phytochemical Screening Using Leaf Extract of Sansevieria Sp. J. Penelit. Pertan. Terap. 2018, 17, 197–202. [Google Scholar] [CrossRef]
- Thu, Z.M.; Oo, S.M.; Nwe, T.M.; Aung, H.T.; Armijos, C.; Hussain, F.H.S.; Vidari, G. Structures and Bioactivities of Steroidal Saponins Isolated from the Genera Dracaena and Sansevieria. Molecules 2021, 26, 1916. [Google Scholar] [CrossRef] [PubMed]
- Danilenko, D.M.; Ring, B.D.; Pierce, G.F. Growth Factors and Cytokines in Hair Follicle Development and Cycling: Recent Insights from Animal Models and the Potentials for Clinical Therapy. Mol. Med. Today 1996, 2, 460–467. [Google Scholar] [CrossRef]
- Kasmawati, H.; Mustarichie, R.; Halimah, E.; Ruslin, R.; A Sida, N. Antialopecia Activity and IR-Spectrometry Characterization of Bioactive Compounds From Sansevieria Trifasciata P. Egypt. J. Chem. 2022, in press. [Google Scholar] [CrossRef]
- Al-Dalahmeh, Y.; Al-Bataineh, N.; Al-Balawi, S.S.; Lahham, J.N.; Al-Momani, I.F.; Al-Sheraideh, M.S.; Mayyas, A.S.; Abu Orabi, S.T.; Al-Qudah, M.A. LC-MS/MS Screening, Total Phenolic, Flavonoid and Antioxidant Contents of Crude Extracts from Three Asclepiadaceae Species Growing in Jordan. Molecules 2022, 27, 859. [Google Scholar] [CrossRef]
- Tribalat, L.; Paisse, O.; Dessalces, G.; Grenier-Loustalot, M.-F. Advantages of LC–MS–MS Compared to LC–MS for the Determination of Nitrofuran Residues in Honey. Anal. Bioanal. Chem. 2006, 386, 2161–2168. [Google Scholar] [CrossRef]
- Shoichet, B.K.; McGovern, S.L.; Wei, B.; Irwin, J.J. Lead Discovery Using Molecular Docking. Curr. Opin. Chem. Biol. 2002, 6, 439–446. [Google Scholar] [CrossRef]
- Arba, M.; Aslikah, N.; Arfan, A.; Ruslin, R.; Yanuar, A. Insight on Estrogen Receptor Alpha Modulator from Indonesian Herbal Database: An in-Silico Analysis. Pharm. J. Farm. Indones. (Pharm. J. Indones.) 2020, 17, 343–351. [Google Scholar] [CrossRef]
- Pan, Z.; Xiong, F.; Chen, Y.-L.; Wan, G.-G.; Zhang, Y.; Chen, Z.-W.; Cao, W.-F.; Zhou, G.-Y. Traceability of Geographical Origin in Gentiana Straminea by UPLC-Q Exactive Mass and Multivariate Analyses. Molecules 2019, 24, 4478. [Google Scholar] [CrossRef] [Green Version]
- Mohamed, S.S.; Fayed, A.H.M. Anti-Obesity Synergistic Effect of Pomegranate Seed Oil (PSO) and Arabic Gum (AG) in Albino Rats. Int. J. Vet. Sci. 2020, 9, 84–89. [Google Scholar]
- Sinaga, N.I.; Hanafi, M.; Yantih, N. Identification Of Chemical Compounds And Antibacterial Activity Of 96% Ethanol Extract From Moringa Oleifera Lam. Leaves Against MRSA (Methicillin Resistant Staphylococcus Aureus). Int. J. Appl. Pharm. 2021, 13, 111–114. [Google Scholar] [CrossRef]
- Shin, H.J.; Lee, H.-S.; Lee, D.-S. The Synergistic Antibacterial Activity of 1-Acetyl-Beta-Carboline and Beta-Lactams against Methicillin-Resistant Staphylococcus Aureus (MRSA). J. Microbiol. Biotechnol. 2010, 20, 501–505. [Google Scholar]
- Lee, H.; Lee, H.; Kwon, Y.; Lee, J.-H.; Kim, J.; Shin, M.-K.; Kim, S.-H.; Bae, H. Methyl Gallate Exhibits Potent Antitumor Activities by Inhibiting Tumor Infiltration of CD4+ CD25+ Regulatory T Cells. J. Immunol. 2010, 185, 6698–6705. [Google Scholar] [CrossRef] [Green Version]
- Sánchez, E.; Heredia, N.; del Camacho-Corona, M.R.; García, S. Isolation, Characterization and Mode of Antimicrobial Action against Vibrio Cholerae of Methyl Gallate Isolated from Acacia Farnesiana. J. Appl. Microbiol. 2013, 115, 1307–1316. [Google Scholar] [CrossRef]
- Kane, C.J.M.; Menna, J.H.; Yeh, Y.-C. Methyl Gallate, Methyl-3,4,5-Trihydroxy-Benzoate, Is a Potent and Highly Specific Inhibitor of Herpes Simplex Virus in Vitro. I. Purification and Characterization of Methyl Gallate from Sapium Sebiferum. Biosci. Rep. 1988, 8, 85–94. [Google Scholar] [CrossRef]
- Al-Omari, S.; Ali, A. Photodynamic Activity of Pyropheophorbide Methyl Ester and Pyropheophorbide a in Dimethylformamide Solution. Gen. Physiol. Biophys. 2009, 28, 70–77. [Google Scholar] [CrossRef] [Green Version]
- Jain, R.; Monthakantirat, O.; Tengamnuay, P.; De-Eknamkul, W. Identification of a New Plant Extract for Androgenic Alopecia Treatment Using a Non-Radioactive Human Hair Dermal Papilla Cell-Based Assay. BMC Complement. Altern. Med. 2016, 16, 18. [Google Scholar] [CrossRef] [Green Version]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [Green Version]
- Oleg, T.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Petrov, D.; Zagrovic, B. Are Current Atomistic Force Fields Accurate Enough to Study Proteins in Crowded Environments? PLoS Comput. Biol. 2014, 10, e1003638. [Google Scholar] [CrossRef] [PubMed]
- Sousa Da Silva, A.W.; Vranken, W.F. ACPYPE—AnteChamber PYthon Parser InterfacE. BMC Res. Notes 2012, 5, 367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Gao, X.; Fang, J. Multiple Staggered Mesh Ewald: Boosting the Accuracy of the Smooth Particle Mesh Ewald Method. J. Chem. Theory Comput. 2016, 12, 5596–5608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasmawati, H.; Mustarichie, R.; Halimah, E.; Ruslin, R.; Arfan, A. The Identification Of Molecular Mechanisms From Bioactive Compounds in Sansevieria Trifasciata Plant as Anti-Alopecia: In-Silico Approach. RASĀYAN J. Chem. 2022, 15, 925–932. [Google Scholar] [CrossRef]
- Pires, D.E.V.; Blundell, T.L.; Ascher, D.B. PkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef]




| Sample | Rt (Min) | Formula | Observed m/z | Neutral Mass (Da) | Identification |
|---|---|---|---|---|---|
| Subfraction C | 5.33 | C13H10N2O | 211.0870 | 210.07931 | 1-Acetyl-ß-carboline |
| 8.82 | C20H20N2O4 | 353.1467 | 352.14231 | Oliveramine | |
| 9.40 | C18H30O2 | 279.2327 | 278.22458 | Trichosanic acid | |
| 10.01 | - | 313.1585 | - | - | |
| 10.19 | C35H36N4O5 | 592.2685 | 593.27650 | Candidate Mass C35H36N4O5 | |
| Subfraction D | 5.81 | C18H18O5 | 315.1232 | 314.11542 | (2S)-3′, 4′-Methylenedioxy-5, 7-dimethoxyflavane |
| 5.81 | C31H27N3O15 | 682.1497 | 681.14422 | Candidate Mass C31H27N3O15 | |
| 10.19 | C35H36N4O5 | 593.2766 | 592.26857 | Candidate Mass C35H36N4O5 | |
| 10.98 | C37H40N4O6 | 637.3024 | 36.294790 | Candidate Mass C37H40N4O6 | |
| 11.57 | C35H38N4O3 | 563.3035 | 562.29439 | Candidate Mass C35H38N4O3 | |
| Subfraction E | 5.80 | C18H18O5 | 315.1230 | 314.11542 | (2S)-3′, 4′-Methylenedioxy-5, 7-dimethoxyflavane |
| 10.63 | C36H38N4O7 | 639.2824 | 638.27405 | Candidate Mass C36H38N4O7 | |
| 10.98 | C36H38N4O5 | 607.2924 | 606.28422 | Candidate Mass C36H38N4O5 | |
| 11.31 | C34H36N4O3 | 549.2870 | 548.27874 | Methyl pyrophaeophorbide A | |
| Subfraction F | 3.37 | C8H8O5 | 185.0438 | 184.03717 | Methyl gallate |
| 3.63 | C11H16O3 | 197.1165 | 196.10994 | Digiprolactone | |
| 9.37 | C18H30O2 | 279.2321 | 278.22458 | Trichosanic acid | |
| 10.16 | C35H36N4O5 | 593.2781 | 592.26857 | Candidate Mass C35H36N4O5 |
| Identified Compounds | Compound’s Code | Docking Score (Kcal/mol) |
|---|---|---|
| Methyl pyrophaeophorbide A | 1 | −7.0 |
| Oliveramine | 2 | −6.3 |
| (2S)-3′, 4′-Methylenedioxy-5, 7-dimethoxyflavane | 3 | −5.8 |
| 1-Acetyl-β-carboline | 4 | −5.2 |
| Digiprolactone | 5 | −4.5 |
| Minoxidil | - | −4.2 |
| Trichosanic acid | 6 | −4.2 |
| Methyl gallate | 7 | −4.0 |
| Compounds | ∆EVDW | ∆EEle | ∆EPS | ∆ESASA | ∆EBind |
|---|---|---|---|---|---|
| Minoxidil | −60.98 | −12.99 | 47.05 | −7.72 | −34.64 |
| 1 | −119.18 | −12.96 | 78.56 | −12.55 | −66.13 |
| 2 | −74.75 | −10.16 | 52.65 | −8.13 | −40.39 |
| 3 | −105.16 | −3.30 | 60.46 | −11.36 | −59.36 |
| 4 | −73.85 | −9.14 | 50.73 | −7.99 | −40.25 |
| Compounds | ADME | Toxicity | ||||||
|---|---|---|---|---|---|---|---|---|
| SP (logKP) | BBB (logBB) | CNS (logPS) | CYP2D6 | TC (mL/min/kg) | AMES | HPT | SS | |
| 1 | −2.854 | 0.038 | −2.611 | No | −0.411 | No | Yes | No |
| 2 | −2.93 | −0.651 | −2.987 | No | 0.765 | Yes | Yes | No |
| 3 | −2.787 | −0.096 | −1.647 | No | 0.214 | No | No | No |
| 4 | −2.85 | 0.584 | −1.39 | No | 0.481 | Yes | No | No |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kasmawati, H.; Mustarichie, R.; Halimah, E.; Ruslin, R.; Arfan, A.; Sida, N.A. Unrevealing the Potential of Sansevieria trifasciata Prain Fraction for the Treatment of Androgenetic Alopecia by Inhibiting Androgen Receptors Based on LC-MS/MS Analysis, and In-Silico Studies. Molecules 2022, 27, 4358. https://doi.org/10.3390/molecules27144358
Kasmawati H, Mustarichie R, Halimah E, Ruslin R, Arfan A, Sida NA. Unrevealing the Potential of Sansevieria trifasciata Prain Fraction for the Treatment of Androgenetic Alopecia by Inhibiting Androgen Receptors Based on LC-MS/MS Analysis, and In-Silico Studies. Molecules. 2022; 27(14):4358. https://doi.org/10.3390/molecules27144358
Chicago/Turabian StyleKasmawati, Henny, Resmi Mustarichie, Eli Halimah, Ruslin Ruslin, Arfan Arfan, and Nurramadhani A. Sida. 2022. "Unrevealing the Potential of Sansevieria trifasciata Prain Fraction for the Treatment of Androgenetic Alopecia by Inhibiting Androgen Receptors Based on LC-MS/MS Analysis, and In-Silico Studies" Molecules 27, no. 14: 4358. https://doi.org/10.3390/molecules27144358