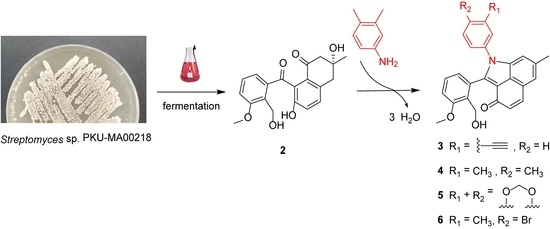
Generation of Unusual Aromatic Polyketides by Incorporation of Phenylamine Analogues into a C-Ring-Cleaved Angucyclinone
Abstract
:1. Introduction
2. Results and Discussion
2.1. Generation of 3–6 by Nonenzymatic Incorporation of Phenylamine Analogues into C-Ring-Cleaved Angucyclinone 2
2.2. Structural Elucidation of 3–6
2.3. Biological Activity Assays
3. Materials and Methods
3.1. General Experimental Procedures
3.2. The Fermentation and Isolation of 2
3.3. The Conversion of 2 to 3–6
3.4. Biological Activity Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kharel, M.K.; Pahari, P.; Shepherd, M.D.; Tibrewal, N.; Nybo, S.E.; Shaaban, K.A.; Rohr, J. Angucyclines: Biosynthesis, mode-of-action, new natural products, and synthesis. Nat. Prod. Rep. 2012, 29, 264–325. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Jin, J.; Yang, X.Y.; Song, J.; Yu, J.H.; Geng, T.T.; Zhang, Z.Y.; Ma, X.Y.; Wang, G.Y.; Xiao, H.; et al. Discovery of a phenylamine-incorporated Angucyclinone from marine Streptomyces sp. PKU-MA00218 and Generation of derivatives with phenylamine analogues. Org. Lett. 2019, 21, 2813–2817. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.J.; Wang, G.Y.; Jin, J.; Liu, T.; Ma, X.Y.; Zhang, Z.Y.; Geng, T.T.; Song, J.; Ma, X.J.; Zhang, Y.T.; et al. Discovery and biosynthesis of pepticinnamins G-M featuring three enzymes-catalyzed nonproteinogenic amino acid formation. J. Org. Chem. 2020, 85, 8673–8682. [Google Scholar] [CrossRef] [PubMed]
- Yuan, B.C.; Liu, D.; Guan, X.; Yan, Y.C.; Zhang, J.P.; Zhang, Y.P.; Yang, D.H.; Ma, M.; Lin, W.H. Piperazine ring formation by a single-module NRPS and cleavage by an α-KG-dependent nonheme iron dioxygenase in brasiliamide biosynthesis. Appl. Microbiol. Biotechnol. 2020, 104, 6149–6159. [Google Scholar] [CrossRef]
- Jin, J.; Yang, X.Y.; Liu, T.; Xiao, H.; Wang, G.Y.; Zhou, M.J.; Liu, F.W.; Zhang, Y.T.; Liu, D.; Chen, M.H.; et al. Fluostatins M-Q featuring a 6-5-6-6 ring skeleton and high oxidized A-rings from marine Streptomyces sp. PKU-MA00045. Mar. Drugs 2018, 16, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, M.J.; Liu, F.W.; Yang, X.Y.; Jin, J.; Dong, X.; Zeng, K.W.; Liu, D.; Zhang, Y.T.; Ma, M.; Yang, D.H. Bacillibactin and bacillomycin analogues with cytotoxicities against human cancer cell lines from marine Bacillus sp. PKU-MA00093 and PKU-MA00092. Mar. Drugs 2018, 16, 22. [Google Scholar] [CrossRef] [Green Version]
- Brusilowskij, B.; Schalley, C.A. Multidentate pyridyl-based ligands in the coordination-driven self-assembly of palladium metallo-macrocycles. Eur. J. Org. Chem. 2011, 2011, 469–477. [Google Scholar] [CrossRef]
- Lai, B.B.; Huang, Z.P.; Jia, Z.F.; Bai, R.X.; Gu, Y.L. Silica supported metal acetylacetonate catalysts with a robust and flexible linker constructed by using butoxy 3,4 dihydropyrans as dual anchoring reagents and ligand donors. Catal. Sci. Technol. 2016, 6, 1810–1820. [Google Scholar] [CrossRef]
- Mukhopadhyay, C.; Tapaswi, P.K. Highly efficient and simple catalytic system for the N-Arylation of some hindered Aza-heterocycles in water. Synth. Commun. 2012, 42, 2217–2228. [Google Scholar] [CrossRef]
- Reem, A.K.A.; Marwa, A.M.S.; Samir, Y.A. Synthesis and anticancer activity of bis-benzo[d][1,3]dioxol-5-yl thiourea derivatives with molecular docking study. Bioorg. Chem. 2019, 90, 103088–103096. [Google Scholar]
- Beresneviiute, K.; Beresnevicius, Z.; Mikulskiene, G.; Kihlberg, J.; Broddefalk, J. 13C NMR study of dihydropyrimidinedione and dihydropyrimidine-2-thione derivatives. Magn. Reson. Chem. 1997, 35, 553–555. [Google Scholar] [CrossRef]
- Liu, Y.X.; Yan, Y.G.; Xue, D.; Wang, Z.F.; Xiao, J.L.; Wang, C. Highly efficient binuclear copper-catalyzed oxidation of N,N-dimethylanilines with O2. ChemCatChem 2020, 12, 2221–2225. [Google Scholar] [CrossRef]
- Hambleton, P.T.; Hedgecock, C.J.R.; Kay, D.P.; Kuo, E.A.; Tully, W.R. Preparation of 2-Cyano-2-(cyclopropylcarbonyl)acetanilides and Analogs as Anti-Inflammatories. Patent PCT EP484223 A2 19920506, 6 May 1992. [Google Scholar]
- Lee, J.M.; Johnson, J.A. An important role of Nrf2-ARE pathway in the cellular defense mechanism. J. Biochem. Mol. Biol. 2004, 37, 139–143. [Google Scholar] [CrossRef] [Green Version]
- Xi, M.Y.; Jia, J.M.; Sun, H.P.; Sun, Z.Y.; Jiang, J.W.; Wang, Y.J.; Zhang, M.Y.; Zhu, J.F.; Xu, L.L.; Jiang, Z.Y.; et al. 3-aroylmethylene-2,3,6,7-tetrahydro-1H-pyrazino[2,1-a]isoquinolin-4(11bH)-ones as potent Nrf2/ARE inducers in human cancer cells and AOM-DSS treated mice. J. Med. Chem. 2013, 56, 7925–7938. [Google Scholar] [CrossRef]
- Ji, S.; Li, R.; Wang, Q.; Miao, W.J.; Li, Z.W.; Si, L.L.; Qiao, X.; Yu, S.W.; Zhou, D.M.; Ye, M. Anti-H1N1 virus, cytotoxic and Nrf2 activation activities of chemical constituents from Scutellaria baicalensis. J. Ethnopharmacol. 2015, 176, 475–484. [Google Scholar] [CrossRef] [PubMed]




| NO. | 1 | 3 | 4 | 5 | 6 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| δH (J in Hz) | δC | δH (J in Hz) | δC | δH (J in Hz) | δC | δH (J in Hz) | δC | δH (J in Hz) | δC | |
| 1 | - | 134.8 | - | 134.9 | - | 135.0 | - | 135.2 | - | 134.8 |
| 2 | 7.27, s | 114.0 | 7.25, s | 113.9 | 7.23, s | 114.2 | 7.26, s | 114.1 | 7.30, s | 114.1 |
| 3 | - | 134.1 | - | 134.3 | - | 134.0 | - | 134.0 | - | 134.3 |
| 4 | 7.56, s | 124.1 | 7.55, s | 124.2 | 7.54, s | 124.1 | 7.54, s | 124.0 | 7.55, s | 124.2 |
| 4a | - | 113.1 | - | 113.1 | - | 113.0 | - | 112.9 | - | 113.1 |
| 5 | 7.85, d (9.6) | 137.2 | 7.85, d (9.6) | 137.3 | 7.84, d (9.5) | 137.2 | 7.84, d (9.5) | 137.2 | 7.84, d (9.5) | 137.3 |
| 6 | 6.53, d (9.6) | 132.9 | 6.52, d (9.6) | 132.9 | 6.52, d (9.5) | 132.9 | 6.51, d (9.5) | 132.9 | 6.52, d (9.5) | 132.9 |
| 6a | - | 180.2 | - | 180.2 | - | 180.2 | - | 180.1 | - | 180.2 |
| 7 | 4.70, d (11.5) | 55.2 | 4.71, br d (9.7) 2 | 55.1 | 4.65, br d (11.0) | 55.4 | 4.71, br d (11.0) | 55.1 | 4.71, br d 2 | 55.1 |
| 4.50, d (11.5) | 4.47, dd (12.1, 7.2) | - | 4.47, br dd (11.0, 4.0) | - | 4.46, br dd (11.0, 4.6) | - | 4.47, dd (12.7, 7.4) | - | ||
| 7a | - | 130.2 | - | 130.3 | - | 130.1 | - | 130.1 | - | 130.2 |
| 8 | - | 157.1 | - | 157.1 | - | 157.2 | - | 157.1 | - | 157.1 |
| 9 | 7.02, dd (8.0, 1.0) | 111.9 | 7.02, d (8.0) | 112.1 | 7.02, d (8.0) | 111.9 | 7.04, d 2 (8.0) | 112.0 | 7.03, d (8.5) | 112.1 |
| 10 | 7.11, t (8.0) | 127.9 | 7.13, t (8.0) | 128.0 | 7.13, t (8.0) | 128.0 | 7.17, t (8.0) | 128.0 | 7.15, t (8.0) | 128.1 |
| 11 | 6.61, dd (8.0, 1.0) | 122.5 | 6.65, br d (7.6) | 122.5 | 6.62, br d (8.0) | 122.5 | 6.67, br d (8.0) | 122.4 | 6.63, br d (7.6) | 122.5 |
| 11a | - | 130.9 | - | 130.8 | - | 131.1 | - | 131.1 | - | 130.9 |
| 12 | - | 145.8 | - | 145.8 | - | 145.9 | - | 146.0 | - | 145.6 |
| 12a | - | 123.0 | - | 123.1 | - | 123.0 | - | 123.0 | - | 123.1 |
| 12b | - | 123.3 | - | 123.3 | - | 123.3 | - | 123.2 | - | 123.3 |
| 13 | 3.85, s | 55.5 | 3.84, s | 55.6 | 3.84, s | 55.6 | 3.85, s | 55.6 | 3.84, s | 55.6 |
| 14 | 2.51, s | 21.5 | 2.51, s 1 | 21.5 | 2.51, s1 | 21.6 | 2.52, s 1 | 21.5 | 2.51, s 1 | 21.5 |
| 7-OH | 4.68, br s | 4.70, br s 2 | - | 4.62, br s | - | 4.69, br s | - | 4.70, br s 2 | - | |
| 1′ | - | 136.0 | - | 136.4 | - | 137.4 | - | 129.7 | - | 135.5 |
| 2′ | 7.54, br d (8.3) | 127.6 | 7.70, br s | 130.7 | 7.36, br s | 128.3 | 7.18, s 2 | 108.2 | 7.61, s 2 | 130.1 |
| 3′ | 7.45, br t (7.7) | 129.3 | - | 122.7 | - | 136.7 | - | 147.1 | - | 138.6 |
| 4′ | 7.41, m | 128.3 | 7.50, d (8.0) | 131.8 | - | 133.6 | - | 147.5 | - | 124.0 |
| 5′ | 7.45, br t (7.7) | 129.3 | 7.45, m | 129.7 | 7.16, br s 2 | 130.1 | 7.04, d 2 | 108.7 | 7.62, d 2 | 132.8 |
| 6′ | 7.54, br d (8.3) | 127.6 | 7.29, m | 128.6 | 7.19, br s 2 | 124.9 | 6.98, br s | 121.6 | 7.28, m | 127.0 |
| 7′ | - | - | 82.4 | 2.28, s | 19.4 | 6.10, br s | 101.9 | 2.32, s | 22.5 | |
| 6.08, br s | ||||||||||
| 8′ | - | 4.31, s | 82.2 | 2.21, s | 19.0 | - | - | - | - | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, H.; Wang, G.; Wang, Z.; Kuang, Y.; Song, J.; Jin, J.; Ye, M.; Yang, D.; Ma, M. Generation of Unusual Aromatic Polyketides by Incorporation of Phenylamine Analogues into a C-Ring-Cleaved Angucyclinone. Molecules 2021, 26, 1959. https://doi.org/10.3390/molecules26071959
Xiao H, Wang G, Wang Z, Kuang Y, Song J, Jin J, Ye M, Yang D, Ma M. Generation of Unusual Aromatic Polyketides by Incorporation of Phenylamine Analogues into a C-Ring-Cleaved Angucyclinone. Molecules. 2021; 26(7):1959. https://doi.org/10.3390/molecules26071959
Chicago/Turabian StyleXiao, Hua, Guiyang Wang, Zhengdong Wang, Yi Kuang, Juan Song, Jing Jin, Min Ye, Donghui Yang, and Ming Ma. 2021. "Generation of Unusual Aromatic Polyketides by Incorporation of Phenylamine Analogues into a C-Ring-Cleaved Angucyclinone" Molecules 26, no. 7: 1959. https://doi.org/10.3390/molecules26071959