Cytotoxic and Antiproliferative Effects of Diarylheptanoids Isolated from Curcuma comosa Rhizomes on Leukaemic Cells
Abstract
:1. Introduction
2. Results and Discussion
2.1. Fractional Extracts and Pure Compounds following Column Chromatography
2.2. Cytotoxicity of Fractional Extracts and Pure Compounds against Leukaemic Cells Compared to Other Cancer Cells Using MTT Assay
2.3. Cytotoxicity of Fractional Extracts and Pure Compounds on PBMCs
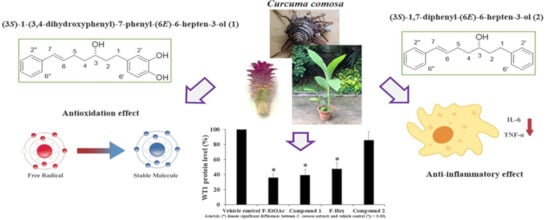
2.4. Antioxidant and Anti-inflammatory Activities of Compound 1 and Compound 2
2.5. Effects of Fractional Extracts and Pure Compounds on Red Blood Cell Haemolysis
2.6. Formatting Effects of Fractional Extracts and Pure Compounds on WT1 Protein Expression and Cell Proliferation
2.7. Effects of Contact Times and Concentrations of Fractional Extracts and Pure Compounds on WT1 Protein Expression and Total Cell Numbers in KG-1a
2.8. Effects of F-EtOAc and Compound 1 on Cell Cycle Distribution in KG-1a Cell Line Using Flow Cytometer
3. Materials and Methods
3.1. Plant Maceration
3.2. Column Chromatography
3.3. Structure Identification
3.4. Cell Culture
3.5. Cytotoxicity Determinations by MTT Assay
3.6. Trypan Blue Exclusion
3.7. Cytotoxicity of PBMCs
3.8. Antioxidant Activities Determination
3.8.1. 2,2′-Azinobis 3-ethylbenzothiazoline-6-sulphonate (ABTS) Assay
3.8.2. 2,2′-Diphenyl-1-picrylhydrazyl-hydrate (DPPH) Assay
3.8.3. Ferric Reducing Antioxidant Power (FRAP) Assay
3.9. Anti-inflammatory Activity Determination
3.10. RBC Haemolysis Induction
3.11. Western Blot Analysis
3.12. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pimkaew, P.; Chuncharunee, A.; Suksamrarn, A.; Piyachaturawat, P. Evaluation on toxicity of Curcuma latifolia Rosc. Thai. J. Toxicol. 2008, 23, 193–196. [Google Scholar]
- Kuttan, R.; Bhanumathy, P.; Nirmala, K.; George, M.C. Potential anticancer activity of turmeric (Curcuma longa). Cancer Lett. 1985, 29, 197–202. [Google Scholar] [PubMed]
- Selvam, R.; Subramanian, L.; Gayathri, R.; Angayarkanni, N. The anti-oxidant activity of turmeric (Curcuma longa). J. Ethnopharmacol. 1995, 47, 59–67. [Google Scholar] [PubMed]
- Menon, V.P.; Sudheer, A.R. Antioxidant and anti-inflammatory properties of curcumin. Adv. Exp. Med. Biol. 2007, 595, 105–125. [Google Scholar]
- Soontornchainaksaeng, P.; Jenjittikul, T. Chromosome number variation of phytoestrogen-producing Curcuma (Zingiberaceae) from Thailand. J. Nat. Med. 2010, 64, 370–377. [Google Scholar]
- Suksamrarn, A.; Ponglikitmongkol, M.; Wongkrajang, K.; Chindaduang, A.; Kittidanairak, S.; Jankam, A.; Yingyongnarongkul, B.-E.; Kittipanumat, N.; Chokchaisiri, R.; Khetkam, P.; et al. Diarylheptanoids, new phytoestrogens from the rhizomes of Curcuma comosa: Isolation, chemical modification and estrogenic activity evaluation. Bioorg. Med. Chem. 2008, 16, 6891–6902. [Google Scholar]
- Tantikanlayaporn, D.; Robinson, L.J.; Suksamrarn, A.; Piyachaturawat, P.; Blair, H. A diarylheptanoid phytoestrogen from Curcuma comosa, 1,7-diphenyl-4,6-heptadien-3-ol, accelerates human osteoblast proliferation and differentiation. Phytomedicine 2013, 20, 676–682. [Google Scholar]
- Jariyawat, S.; Thammapratip, T.; Suksen, K.; Wanitchakool, P.; Nateewattana, J.; Chairoungdua, A.; Suksamrarn, A.; Piyachaturawat, P. Induction of apoptosis in murine leukemia by diarylheptanoids from Curcuma comosa Roxb. Cell Biol. Toxicol. 2011, 27, 413–423. [Google Scholar]
- Anuchapreeda, S.; Rungrojsakul, M.; Tima, S.; Chiampanichayakul, S.; Krig, S.R. Co-activation of WT1 and AP-1 proteins on WT1 gene promoter to induce WT1 gene expression in K562 cells. Cell. Signal. 2019, 53, 339–347. [Google Scholar]
- Suksamrarn, A.; Eiamong, S.; Piyachaturawat, P.; Charoenpiboonsin, J. Phenolic diarylheptanoids from Curcuma xanthorrhiza. Phytochemistry 1994, 36, 1505–1508. [Google Scholar]
- Li, J.; Zhao, F.; Li, M.Z.; Chen, L.X.; Qiu, F. Diarylheptanoids from the Rhizomes of Curcuma kwangsiensis. J. Nat. Prod. 2010, 73, 1667–1671. [Google Scholar] [PubMed]
- Kuroyanagi, M.; Noro, T.; Fukushima, S.; Aiyama, R.; Ikuta, A.; Itokawa, H.; Morita, M. Studies on the constituents of the seeds of Alpinia katsumadai Hayata. Chem. Pharm. Bull. 1983, 31, 1544–1550. [Google Scholar]
- Jurgens, T.M.; Frazier, E.G.; Schaeffer, J.M.; Jones, T.E.; Zink, D.L.; Borris, R.P.; Nanakorn, W.; Beck, H.T.; Balick, M.J. Novel nematocidal agents from Curcuma comosa. J. Nat. Prod. 1994, 57, 230–235. [Google Scholar] [PubMed]
- Rodrigues, M.D.D.; Santiago, P.; Marques, K.M.R.; Pereira, V.R.A.; de Castro, M.; Cantalice, J.; da Silva, T.G.; Adam, M.L.; do Nascimento, S.C.; de Albuquerque, J.F.C.; et al. Selective cytotoxic and genotoxic activities of 5-(2-bromo-5-methoxybenzylidene)-thiazolidine-2,4-dione against NCI-H292 human lung carcinoma cells. Pharmacol. Rep. 2018, 70, 446–454. [Google Scholar] [PubMed]
- Griffiths, H.R.; Lunec, J. Ascorbic acid in the 21st century—More than a simple antioxidant. Environ. Toxicol. Pharmacol. 2001, 10, 173–182. [Google Scholar] [PubMed]
- Jitsanong, T.; Khanobdee, K.; Piyachaturawat, P.; Wongprasert, K. Diarylheptanoid 7-(3,4 dihydroxyphenyl)-5-hydroxy-1-phenyl-(1E)-1-heptene from Curcuma comosa Roxb. protects retinal pigment epithelial cells against oxidative stress-induced cell death. Toxicol. In Vitro 2011, 25, 167–176. [Google Scholar]
- Krishnaiah, D.; Sarbatly, R.; Nithyanandam, R. A review of the antioxidant potential of medicinal plant species. Food Bioprod. Process. 2011, 89, 217–233. [Google Scholar]
- Uttara, B.; Singh, A.V.; Zamboni, P.; Mahajan, R.T. Oxidative stress and neurodegenerative diseases: A review of upstream and downstream antioxidant therapeutic options. Curr. Neuropharmacol. 2009, 7, 65–74. [Google Scholar]
- Bjelakovic, G.; Nikolova, D.; Simonetti, R.G.; Gluud, C. Antioxidant supplements for prevention of gastrointestinal cancers: A systematic review and meta-analysis. Lancet 2004, 364, 1219–1228. [Google Scholar]
- Schubert, M.A.; Müller-Goymann, C.C. Characterisation of surface-modified solid lipid nanoparticles (SLN): Influence of lecithin and nonionic emulsifier. Eur. J. Pharm. Biopharm. 2005, 61, 77–86. [Google Scholar]
- Panyajai, P.; Amnajphook, N.; Keawsangthongcharoen, S.; Chiampanichayakul, S.; Tima, S.; Anuchapreeda, S. Study of leukemic stem cell population (CD34+/CD38-) and WT1 protein expression in human leukemic cell lines. J. Assoc. Med. Sci. 2018, 51, 38–44. [Google Scholar]
- Anuchapreeda, S.; Khumpirapang, N.; Rupitiwiriya, K.; Tho-Iam, L.; Saiai, A.; Okonogi, S.; Usuki, T. Cytotoxicity and inhibition of leukemic cell proliferation by sesquiterpenes from rhizomes of Mah-Lueang (Curcuma cf. viridiflora Roxb.). Bioorg. Med. Chem. Lett. 2018, 28, 410–414. [Google Scholar] [PubMed]
- Chaiyana, W.; Punyoyai, C.; Somwongin, S.; Leelapornpisid, P.; Ingkaninan, K.; Waranuch, N.; Srivilai, J.; Thitipramote, N.; Wisuitiprot, W.; Schuster, R.; et al. Inhibition of 5alpha-Reductase, IL-6 Secretion, and Oxidation Process of Equisetum debile Roxb. ex Vaucher Extract as Functional Food and Nutraceuticals Ingredients. Nutrients 2017, 9, 1105. [Google Scholar]
- Chaiyana, W.; Anuchapreeda, S.; Punyoyai, C.; Neimkhum, W.; Lee, K.-H.; Lin, W.-C.; Lue, S.-C.; Viernstein, H.; Mueller, M. Ocimum sanctum Linn. as a natural source of skin anti-ageing compounds. Ind. Crop. Prod. 2019, 127, 217–224. [Google Scholar]











| Samples | TEAC (μM Torox/g Extract) | EC1 (mM FeSo4/g Extract) | IC50 DPPH (μg/mL) |
|---|---|---|---|
| Ascorbic acid | 1.7 ± 0.2 a | 23.7 ± 0.7 a | 13.9 ± 0.5 a |
| Compound 1 | 0.9 ± 0.2 a | 23.2 ± 1.0 a | 13.0 ± 0.3 b |
| Compound 2 | 0.1 ± 0.0 b | 1.5 ± 0.0 b | >100 c |
Samples Availability: Samples of the compounds not available. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Viriyaadhammaa, N.; Saiai, A.; Neimkhum, W.; Nirachonkul, W.; Chaiyana, W.; Chiampanichayakul, S.; Tima, S.; Usuki, T.; Duangmano, S.; Anuchapreeda, S. Cytotoxic and Antiproliferative Effects of Diarylheptanoids Isolated from Curcuma comosa Rhizomes on Leukaemic Cells. Molecules 2020, 25, 5476. https://doi.org/10.3390/molecules25225476
Viriyaadhammaa N, Saiai A, Neimkhum W, Nirachonkul W, Chaiyana W, Chiampanichayakul S, Tima S, Usuki T, Duangmano S, Anuchapreeda S. Cytotoxic and Antiproliferative Effects of Diarylheptanoids Isolated from Curcuma comosa Rhizomes on Leukaemic Cells. Molecules. 2020; 25(22):5476. https://doi.org/10.3390/molecules25225476
Chicago/Turabian StyleViriyaadhammaa, Natsima, Aroonchai Saiai, Waranya Neimkhum, Wariya Nirachonkul, Wantida Chaiyana, Sawitree Chiampanichayakul, Singkome Tima, Toyonobu Usuki, Suwit Duangmano, and Songyot Anuchapreeda. 2020. "Cytotoxic and Antiproliferative Effects of Diarylheptanoids Isolated from Curcuma comosa Rhizomes on Leukaemic Cells" Molecules 25, no. 22: 5476. https://doi.org/10.3390/molecules25225476