Protein-Assisted Room-Temperature Assembly of Rigid, Immobile Holliday Junctions and Hierarchical DNA Nanostructures
Abstract
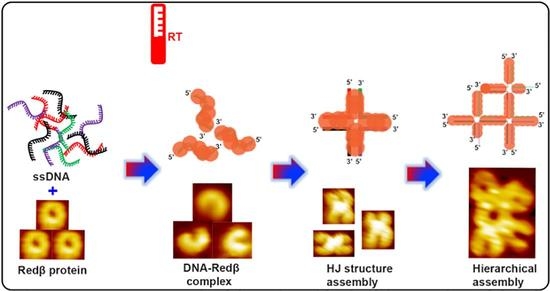
:1. Introduction
2. Results and Discussion
2.1. Assembly of Blunt-End HJ Structures from 44 mer, 66 mer, and 88 mer Oligonucleotides at 37 °C
2.2. Hierarchical Assembly of HJ Structures with Sticky Ends
2.3. Assembly of Hierarchical Networks Composed of 66 mer HJ Structures
3. Materials and Methods
3.1. Protein Expression, Affinity, and Preparative Gel Filtration Chromatography
3.2. Establishment of a Holliday Junction Assembly Protocol Using Redβ
3.3. Redβ-Mediated Annealing and Purification of Hierarchical HJ Assemblies with Sticky Ends (S1S4 and S5S8)
3.4. Redβ-Mediated Assembly of Two-Dimensional Hierarchical HJ Networks (S9S12)
3.5. AFM Imaging
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Brázda, V.; Laister, R.C.; Jagelská, E.B.; Arrowsmith, C. Cruciform structures are a common DNA feature important for regulating biological processes. BMC Mol. Biol. 2011, 12, 33. [Google Scholar] [CrossRef] [Green Version]
- Eichman, B.F.; Ortiz-Lombardı́a, M.; Aymamı́, J.; Coll, M.; Ho, P.S. The Inherent Properties of DNA Four-way Junctions: Comparing the Crystal Structures of Holliday Junctions. J. Mol. Biol. 2002, 320, 1037–1051. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.; Hong, S.; Guan, Z.; He, W.; Zhang, D.; Yin, P. Structural insights into sequence-dependent Holliday junction resolution by the chloroplast resolvase MOC1. Nat. Commun. 2020, 11, 1417. [Google Scholar] [CrossRef]
- Mount, A.R.; Mountford, C.P.; Evans, S.A.G.; Su, T.-J.; Buck, A.H.; Dickinson, P.; Campbell, C.J.; Keane, L.M.; Terry, J.G.; Beattie, J.S.; et al. The stability and characteristics of a DNA Holliday junction switch. Biophys. Chem. 2006, 124, 214–221. [Google Scholar] [CrossRef]
- Panyutin, I.G.; Biswas, I.; Hsieh, P. A pivotal role for the structure of the Holliday junction in DNA branch migration. EMBO J. 1995, 14, 1819–1826. [Google Scholar] [CrossRef] [PubMed]
- Panyutin, I.G.; Hsieh, P. The kinetics of spontaneous DNA branch migration. Proc. Natl. Acad. Sci. USA 1994, 91, 2021–2025. [Google Scholar] [CrossRef] [Green Version]
- Komori, K.; Sakae, S.; Shinagawa, H.; Morikawa, K.; Ishino, Y. A Holliday junction resolvase from Pyrococcus furiosus: Functional similarity to Escherichia coli RuvC provides evidence for conserved mechanism of homologous recombination in Bacteria, Eukarya, and Archaea. Proc. Natl. Acad. Sci. USA 1999, 96, 8873–8878. [Google Scholar] [CrossRef] [Green Version]
- Seeman, N.C. Biochemistry and structural DNA nanotechnology: An evolving symbiotic relationship. Biochemistry 2003, 42, 7259–7269. [Google Scholar] [CrossRef]
- Seeman, N.C. Nucleic acid junctions and lattices. J. Theor. Biol. 1982, 99, 237–247. [Google Scholar] [CrossRef]
- Lu, M.; Guo, Q.; Mueller, J.E.; Kemper, B.; Studier, F.W.; Seeman, N.C.; Kallenbach, N.R. Characterization of a bimobile DNA junction. J. Biol. Chem. 1990, 265, 16778–16785. [Google Scholar]
- Marky, L.A.; Kallenbach, N.R.; McDonough, K.A.; Seeman, N.C.; Breslauer, K.J. The melting behavior of a DNA junction structure: A calorimetric and spectroscopic study. Biopolymers 1987, 26, 1621–1634. [Google Scholar] [CrossRef]
- Chen, J.H.; Churchill, M.E.; Tullius, T.D.; Kallenbach, N.R.; Seeman, N.C. Construction and analysis of monomobile DNA junctions. Biochemistry 1988, 27, 6032–6038. [Google Scholar] [CrossRef]
- Seeman, N.C.; Kallenbach, N.R. Design of immobile nucleic acid junctions. Biophys. J. 1983, 44, 201–209. [Google Scholar] [CrossRef] [Green Version]
- Wemmer, D.E.; Wand, A.J.; Seeman, N.C.; Kallenbach, N.R. NMR analysis of DNA junctions: Imino proton NMR studies of individual arms and intact junction. Biochemistry 1985, 24, 5745–5749. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.Q.; Kan, M.K.; Sato, G.H.; Okamoto, T.; Sato, J.D. Characterization and molecular cloning of a putative binding protein for heparin-binding growth factors. J. Biol. Chem. 1991, 266, 16778–16785. [Google Scholar] [PubMed]
- Mikheikin, A.L.; Lushnikov, A.Y.; Lyubchenko, Y.L. Effect of DNA supercoiling on the geometry of holliday junctions. Biochemistry 2006, 45, 12998–13006. [Google Scholar] [CrossRef] [Green Version]
- Vander Zanden, C.M.; Rowe, R.K.; Broad, A.J.; Robertson, A.B.; Ho, P.S. Effect of Hydroxymethylcytosine on the Structure and Stability of Holliday Junctions. Biochemistry 2016, 55, 5781–5789. [Google Scholar] [CrossRef] [Green Version]
- Ho, P.S. Structure of the Holliday junction: Applications beyond recombination. Biochem. Soc. Trans. 2017, 45, 1149–1158. [Google Scholar] [CrossRef]
- Zheng, J.; Birktoft, J.J.; Chen, Y.; Wang, T.; Sha, R.; Constantinou, P.E.; Ginell, S.L.; Mao, C.; Seeman, N.C. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature 2009, 461, 74–77. [Google Scholar] [CrossRef]
- Bergen, A.; Rudiuk, S.; Morel, M.; Le Saux, T.; Ihmels, H.; Baigl, D. Photodependent Melting of Unmodified DNA Using a Photosensitive Intercalator: A New and Generic Tool for Photoreversible Assembly of DNA Nanostructures at Constant Temperature. Nano Lett. 2016, 16, 773–780. [Google Scholar] [CrossRef]
- Fu, Y.; Zeng, D.; Chao, J.; Jin, Y.; Zhang, Z.; Liu, H.; Di, L.; Ma, H.; Huang, Q.; Gothelf, K.V.; et al. Single-step rapid assembly of DNA origami nanostructures for addressable nanoscale bioreactors. J. Am. Chem. Soc. 2013, 135, 696–702. [Google Scholar] [CrossRef] [PubMed]
- Jungmann, R.; Liedl, T.; Sobey, T.L.; Shih, W.; Simmel, F.C. Isothermal assembly of DNA origami structures using denaturing agents. J. Am. Chem. Soc. 2008, 130, 10062–10063. [Google Scholar] [CrossRef]
- Kopielski, A.; Schneider, A.; Csáki, A.; Fritzsche, W. Isothermal DNA origami folding: Avoiding denaturing conditions for one-pot, hybrid-component annealing. Nanoscale 2015, 7, 2102–2106. [Google Scholar] [CrossRef] [PubMed]
- Myhrvold, C.; Dai, M.; Silver, P.A.; Yin, P. Isothermal self-assembly of complex DNA structures under diverse and biocompatible conditions. Nano Lett. 2013, 13, 4242–4248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sobczak, J.-P.J.; Martin, T.G.; Gerling, T.; Dietz, H. Rapid folding of DNA into nanoscale shapes at constant temperature. Science 2012, 338, 1458–1461. [Google Scholar] [CrossRef]
- Wang, D.; Liu, Q.; Wu, D.; He, B.; Li, J.; Mao, C.; Wang, G.; Qian, H. Isothermal Self-Assembly of Spermidine-DNA Nanostructure Complex as a Functional Platform for Cancer Therapy. ACS Appl. Mater. Interfaces 2018, 10, 15504–15516. [Google Scholar] [CrossRef]
- Caldwell, B.J.; Bell, C.E. Structure and mechanism of the Red recombination system of bacteriophage λ. Prog. Biophys. Mol. Biol. 2019, 147, 33–46. [Google Scholar] [CrossRef]
- Kuzminov, A. Homologous Recombination-Experimental Systems, Analysis, and Significance. EcoSal Plus 2011, 4. [Google Scholar] [CrossRef] [Green Version]
- Erler, A.; Wegmann, S.; Elie-Caille, C.; Bradshaw, C.R.; Maresca, M.; Seidel, R.; Habermann, B.; Muller, D.J.; Stewart, A.F. Conformational adaptability of Redbeta during DNA annealing and implications for its structural relationship with Rad52. J. Mol. Biol. 2009, 391, 586–598. [Google Scholar] [CrossRef]
- Muyrers, J.P.P.; Zhang, Y.; Stewart, A.F. Techniques: Recombinogenic engineering–new options for cloning and manipulating DNA. Trends Biochem. Sci. 2001, 26, 325–331. [Google Scholar] [CrossRef]
- Copeland, N.G.; Jenkins, N.A.; Court, D.L. Recombineering: A powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2001, 2, 769–779. [Google Scholar] [CrossRef]
- Zhang, Y.; Buchholz, F.; Muyrers, J.P.; Stewart, A.F. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 1998, 20, 123–128. [Google Scholar] [CrossRef]
- Ander, M.; Subramaniam, S.; Fahmy, K.; Stewart, A.F.; Schäffer, E. A Single-Strand Annealing Protein Clamps DNA to Detect and Secure Homology. PLoS Biol. 2015, 13, e1002213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramaniam, S.; Erler, A.; Fu, J.; Kranz, A.; Tang, J.; Gopalswamy, M.; Ramakrishnan, S.; Keller, A.; Grundmeier, G.; Müller, D.; et al. DNA annealing by Redβ is insufficient for homologous recombination and the additional requirements involve intra- and inter-molecular interactions. Sci. Rep. 2016, 6, 34525. [Google Scholar] [CrossRef] [PubMed]
- Teschome, B.; Facsko, S.; Schönherr, T.; Kerbusch, J.; Keller, A.; Erbe, A. Temperature-Dependent Charge Transport through Individually Contacted DNA Origami-Based Au Nanowires. Langmuir 2016, 32, 10159–10165. [Google Scholar] [CrossRef] [PubMed]
- Aryal, B.R.; Ranasinghe, D.R.; Westover, T.R.; Calvopiña, D.G.; Davis, R.C.; Harb, J.N.; Woolley, A.T. DNA origami mediated electrically connected metal—Semiconductor junctions. Nano Res. 2020, 13, 1419–1426. [Google Scholar] [CrossRef]
- Shen, B.; Linko, V.; Tapio, K.; Pikker, S.; Lemma, T.; Gopinath, A.; Gothelf, K.V.; Kostiainen, M.A.; Toppari, J.J. Plasmonic nanostructures through DNA-assisted lithography. Sci. Adv. 2018, 4, eaap8978. [Google Scholar] [CrossRef] [Green Version]
- Urban, M.J.; Dutta, P.K.; Wang, P.; Duan, X.; Shen, X.; Ding, B.; Ke, Y.; Liu, N. Plasmonic Toroidal Metamolecules Assembled by DNA Origami. J. Am. Chem. Soc. 2016, 138, 5495–5498. [Google Scholar] [CrossRef] [Green Version]
- Ramakrishnan, S.; Subramaniam, S.; Stewart, A.F.; Grundmeier, G.; Keller, A. Regular Nanoscale Protein Patterns via Directed Adsorption through Self-Assembled DNA Origami Masks. ACS Appl. Mater. Interfaces 2016, 8, 31239–31247. [Google Scholar] [CrossRef]
- Liu, L.; Zheng, M.; Li, Z.; Li, Q.; Mao, C. Patterning Nanoparticles with DNA Molds. ACS Appl. Mater. Interfaces 2019, 11, 13853–13858. [Google Scholar] [CrossRef]
- Zhu, J.; Wei, B.; Yuan, Y.; Mi, Y. UNIQUIMER 3D, a software system for structural DNA nanotechnology design, analysis and evaluation. Nucleic Acids. Res. 2009, 37, 2164–2175. [Google Scholar] [CrossRef] [Green Version]
- Praetorius, F.; Dietz, H. Self-assembly of genetically encoded DNA-protein hybrid nanoscale shapes. Science 2017, 355. [Google Scholar] [CrossRef]
- Schiffels, D.; Szalai, V.A.; Liddle, J.A. Molecular Precision at Micrometer Length Scales: Hierarchical Assembly of DNA-Protein Nanostructures. ACS Nano 2017, 11, 6623–6629. [Google Scholar] [CrossRef]
- Lilley, D.M. Structures of helical junctions in nucleic acids. Q. Rev. Biophys. 2000, 33, 109–159. [Google Scholar] [CrossRef]
- McKinney, S.A.; Freeman, A.D.J.; Lilley, D.M.J.; Ha, T. Observing spontaneous branch migration of Holliday junctions one step at a time. Proc. Natl. Acad. Sci. USA 2005, 102, 5715–5720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skerra, A. Use of the tetracycline promoter for the tightly regulated production of a murine antibody fragment in Escherichia coli. Gene 1994, 151, 131–135. [Google Scholar] [CrossRef]








Sample Availability: Samples of the compounds are not available from the authors. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramakrishnan, S.; Subramaniam, S.; Kielar, C.; Grundmeier, G.; Stewart, A.F.; Keller, A. Protein-Assisted Room-Temperature Assembly of Rigid, Immobile Holliday Junctions and Hierarchical DNA Nanostructures. Molecules 2020, 25, 5099. https://doi.org/10.3390/molecules25215099
Ramakrishnan S, Subramaniam S, Kielar C, Grundmeier G, Stewart AF, Keller A. Protein-Assisted Room-Temperature Assembly of Rigid, Immobile Holliday Junctions and Hierarchical DNA Nanostructures. Molecules. 2020; 25(21):5099. https://doi.org/10.3390/molecules25215099
Chicago/Turabian StyleRamakrishnan, Saminathan, Sivaraman Subramaniam, Charlotte Kielar, Guido Grundmeier, A. Francis Stewart, and Adrian Keller. 2020. "Protein-Assisted Room-Temperature Assembly of Rigid, Immobile Holliday Junctions and Hierarchical DNA Nanostructures" Molecules 25, no. 21: 5099. https://doi.org/10.3390/molecules25215099