Modified Nucleic Acids: Expanding the Capabilities of Functional Oligonucleotides
Abstract
:1. General Introduction
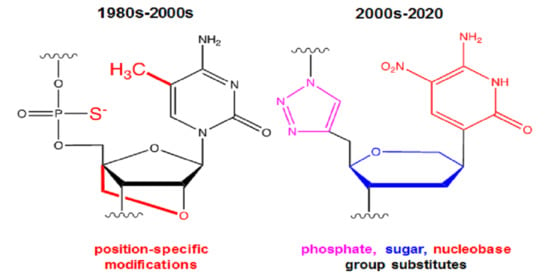
1.1. Overview of Historically Popular Backbone Chemical Modifications Intended to Enhance In Vivo Performance of Modified Oligonucleotides Acting as ASO or Aptamer Sequences
1.2. Chemical Substitutions and Bridges in Pentose Sugar Groups
1.3. Phosphodiester Linkage Modifications and Complete Backbone Replacement Strategies
1.4. Structural Modification Involving Mirror Image Analogs to Natural d-Oligonucleotides
1.5. Performance of Modified Oligonucleotides as Primary Hybridization Partners for Antisense Therapeutics and Gene Editing
1.6. Performance of Modified Oligonucleotides in Double-Stranded Probe Systems for Displacement Strategies
1.7. Current Limitations of Popular Chemical Substitutions and Preview of Recent Approaches Involving Artificial Chemical Groups
2. More Recent Efforts to Expand Oligonucleotide Modification Approaches to Enhance Their Functionality
2.1. Artificial Sugar Groups in Xeno-Nucleic Acids (XNA)
2.2. Modifying Nucleobases with Singular Hydrophobic Groups, Chemical Handles or Carbohydrates
2.3. Xenobiotic Nucleobases as Artificial Base-Pair Matches
2.4. Further Expanding Library Diversity Using Multiple Modifications “Coded” into DNA Sequence
2.5. Various Modified Aptamers as Protein Activity Regulators
3. Modified Nucleic Acid Enzymes
3.1. Modified Nucleic Acid Enzymes with Efficient and Novel Catalytic Activity
3.2. Spatiotemporally Controlled Synthetic Catalysts
3.3. Future Outlook
Funding
Conflicts of Interest
Abbreviations
| ASO—antisense oligonucleotides |
| CeNA—cyclohexene nucleic acids (sugar modification) |
| FANA—2′-fluoroarabino nucleic acids (sugar modification) |
| HNA—hexitol nucleic acid |
| LIVE—laboratory in vitro evolution |
| LNA—locked nucleic acid |
| LOOPER—ligase-catalyzed oligonucleotide polymerization |
| NANP—nucleic acid nanoparticles |
| PCR—polymerase chain reaction |
| PEG—polyethylene glycol |
| phNA—P-methyl/ethyl-phosphonate |
| PNA—peptide nucleic acid |
| PS—phosphorothioate |
| SELEX—systematic evolution of ligands by exponential enrichment |
| SPS—solid phase synthesis |
| TNA—threose nucleic acid |
| VEGF—vascular endothelial growth factor |
| XNA—xeno nucleic acids |
| XNAzymes—xeno nucleic acid enzymes |
References
- Zamecnik, P.C.; Stephenson, M.L. Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc. Natl. Acad. Sci. USA 1978, 75, 280–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, S.; Yao, H.; Wang, L.; Lu, J.; Jiang, F.; Lu, A.; Zhang, G. Chemical modifications of nucleic acid aptamers for therapeutic purposes. Int. J. Mol. Sci. 2017, 18, 1683. [Google Scholar] [CrossRef] [PubMed]
- Robertson, D.L.; Joyce, G.F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 1990, 344, 467–468. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Goodchild, J. Conjugates of oligonucleotides and modified oligonucleotides: A review of their synthesis and properties. Bioconjug. Chem. 1990, 1, 165–187. [Google Scholar] [CrossRef]
- Freier, S.M.; Altmann, K.H. The ups and downs of nucleic acid duplex stability: Structure-stability studies on chemically-modified DNA: RNA duplexes. Nucleic Acids Res. 1997, 25, 4429–4443. [Google Scholar] [CrossRef] [Green Version]
- Pinheiro, V.B.; Taylor, A.I.; Cozens, C.; Abramov, M.; Renders, M.; Zhang, S.; Chaput, J.C.; Wengel, J.; Peak-Chew, S.Y.; McLaughlin, S.H.; et al. Synthetic genetic polymers capable of heredity and evolution. Science 2012, 336, 341–344. [Google Scholar] [CrossRef] [Green Version]
- Dellafiore, M.A.; Montserrat, J.M.; Iribarren, A.M. Modified nucleoside triphosphates for in vitro selection techniques. Front. Chem. 2016, 4, 18. [Google Scholar] [CrossRef] [Green Version]
- Friedman, A.D.; Kim, D.; Li, R. Highly stable aptamers selected from a 2′-fully modified fGmH RNA library for targeting biomaterials. Biomaterials 2015, 36, 110–123. [Google Scholar] [CrossRef] [Green Version]
- Petersen, M.; Nielsen, C.B.; Nielsen, K.E.; Jensen, G.A.; Bondensgaard, K.; Singh, S.K.; Rajwanshi, V.K.; Koshkin, A.A.; Dahl, B.M.; Wengel, J.; et al. The conformations of locked nucleic acids (LNA). J. Mol. Recogn. 2000, 13, 44–53. [Google Scholar] [CrossRef]
- Eremeeva, E.; Fikatas, A.; Margamuljana, L.; Abramov, M.; Schols, D.; Groaz, E.; Herdewijn, P. Highly stable hexitol based XNA aptamers targeting the vascular endothelial growth factor. Nucleic Acids Res. 2019, 47, 4927–4939. [Google Scholar] [CrossRef]
- Sharma, V.K.; Sharma, R.K.; Singh, S.K. Antisense oligonucleotides: Modifications and clinical trials. Med. Chem. Commun. 2014, 5, 1454–1471. [Google Scholar] [CrossRef]
- Varizhuk, A.M.; Kaluzhny, D.N.; Novikov, R.A.; Chizhov, A.O.; Smirnov, I.P.; Chuvilin, A.N.; Tatarinova, O.N.; Fisunov, G.Y.; Pozmogova, G.E.; Florentiev, V.L. Synthesis of triazole-linked oligonucleotides with high affinity to DNA complements and an analysis of their compatibility with biosystems. J. Org. Chem. 2013, 78, 5964–5969. [Google Scholar] [CrossRef] [PubMed]
- Frey, P.A.; Sammons, R.D. Bond order and charge localization in nucleoside phosphorothioates. Science 1985, 228, 541–545. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Allen, L.C. Sulfur does not form double bonds in phosphorothioate anions. J. Am. Chem. Soc. 1987, 109, 6449–6453. [Google Scholar] [CrossRef]
- Ruckman, J.; Green, L.S.; Beeson, J.; Waugh, S.; Gillette, W.L.; Henninger, D.D.; Claesson-Welsh, L.; Janjić, N. 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165): Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem. 1998, 273, 20556–20567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinheiro, V.B.; Holliger, P. Towards XNA nanotechnology: New materials from synthetic genetic polymers. Trends Biotechnol. 2014, 32, 321–328. [Google Scholar] [CrossRef] [Green Version]
- Hermans, N.; Huisman, J.J.; Brouwer, T.B.; Schächner, C.; van Heusden, G.P.H.; Griesenbeck, J.; van Noort, J. Toehold-enhanced LNA probes for selective pull down and single-molecule analysis of native chromatin. Sci. Rep. 2017, 7, 16721. [Google Scholar] [CrossRef]
- Peel, B.J.; Hagen, G.; Krishnamurthy, K.; Desaulniers, J.P. Conjugation and evaluation of small hydrophobic molecules to triazole-linked siRNAs. ACS Med. Chem. Lett. 2015, 6, 117–122. [Google Scholar] [CrossRef] [Green Version]
- Venkiteswaran, S.; Vijayanathan, V.; Shirahata, A.; Thomas, T.; Thomas, T.J. Antisense recognition of the HER-2 mRNA: Effects of phosphorothioate substitution and polyamines on DNA.RNA, RNA.RNA, and DNA.DNA duplex stability. Biochemistry 2005, 44, 303–312. [Google Scholar] [CrossRef] [PubMed]
- King, D.J.; Ventura, D.A.; Brasier, A.R.; Gorenstein, D.G. Novel combinatorial selection of phosphorothioate oligonucleotide aptamers. Biochemistry 1998, 37, 16489–16493. [Google Scholar] [CrossRef] [PubMed]
- Arangundy-Franklin, S.; Taylor, A.I.; Porebski, B.T.; Genna, V.; Peak-Chew, S.; Vaisman, A.; Woodgate, R.; Orozco, M.; Holliger, P. A synthetic genetic polymer with an uncharged backbone chemistry based on alkyl phosphonate nucleic acids. Nat. Chem. 2019, 11, 533–542. [Google Scholar] [CrossRef]
- Buhr, C.A.; Wagner, R.W.; Grant, D.; Froehler, B.C. Oligodeoxynucleotides containing C-7 propyne analogs of 7-deaza-2′-deoxyguanosine and 7-deaza-2′-deoxyadenosine. Nucleic Acids Res. 1996, 24, 2974–2980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheng, P.; Yang, Z.; Kim, Y.; Wu, Y.; Tan, W.; Benner, S.A. Design of a novel molecular beacon: Modification of the stem with artificially genetic alphabet. Chem. Commun. 2008, 5128–5130. [Google Scholar] [CrossRef] [PubMed]
- Kimoto, M.; Hirao, I. Unique thermal stability of unnatural hydrophobic Ds bases in double-stranded DNAs. ACS Synth. Biol. 2017, 6, 1944–1951. [Google Scholar] [CrossRef] [PubMed]
- Wolk, S.K.; Shoemaker, R.K.; Mayfield, W.S.; Mestdagh, A.L.; Janjic, N. Influence of 5-N-carboxamide modifications on the thermodynamic stability of oligonucleotides. Nucleic Acids Res. 2015, 43, 9107–9122. [Google Scholar] [CrossRef] [PubMed]
- Maio, G.; Enweronye, O.; Zumrut, H.E.; Batool, S.; Van, N.; Mallikaratchy, P. Systematic optimization and modification of a DNA aptamer with 2′-O-methyl RNA analogues. ChemistrySelect 2017, 2, 2335–2340. [Google Scholar] [CrossRef] [Green Version]
- Ng, E.W.; Shima, D.T.; Calias, P.; Cunningham, E.T., Jr.; Guyer, D.R.; Adamis, A.P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug. Discov. 2006, 5, 123–132. [Google Scholar] [CrossRef]
- Mi, J.; Liu, Y.; Rabbani, Z.N.; Yang, Z.; Urban, J.H.; Sullenger, B.A.; Clary, B.M. In vivo selection of tumor-targeting RNA motifs. Nat. Chem. Biol. 2010, 6, 22–24. [Google Scholar] [CrossRef]
- Mi, J.; Ray, P.; Liu, J.; Kuan, C.T.; Xu, J.; Hsu, D.; Sullenger, B.A.; White, R.R.; Clary, B.M. Selection against human colorectal cancer xenografts identifies an aptamer that targets RNA helicase protein DHX9. Mol. Ther. Nucleic Acids 2016, 5, e315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Martino, M.T.; Gulla, A.; Gallo Cantafio, M.E.; Altomare, E.; Amodio, N.; Leone, E.; Morelli, E.; Lio, S.G.; Caracciolo, D.; Rossi, M.; et al. In vitro and in vivo activity of a novel locked nucleic acid (LNA)-inhibitor-miR-221 against multiple myeloma cells. PLoS ONE 2014, 9, e89659. [Google Scholar] [CrossRef]
- Kurreck, J.; Wyszko, E.; Gillen, C.; Erdmann, V.A. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 2002, 30, 1911–1918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eze, N.A.; Milam, V.T. Exploring locked nucleic acids as a bio-inspired materials assembly and disassembly tool. Soft Matter 2013, 9, 2403–2411. [Google Scholar] [CrossRef]
- Eze, N.A.; Sullivan, R.S.; Milam, V.T. Analysis of in situ LNA and DNA hybridization events on microspheres. Biomacromolecules 2017, 18, 1086–1096. [Google Scholar] [CrossRef]
- Veedu, R.N.; Vester, B.; Wengel, J. Efficient enzymatic synthesis of LNA-modified DNA duplexes using KOD DNA polymerase. Org. Biomol. Chem. 2009, 7, 1404–1409. [Google Scholar] [CrossRef]
- Shi, H.; He, X.; Cui, W.; Wang, K.; Deng, K.; Li, D.; Xu, F. Locked nucleic acid/DNA chimeric aptamer probe for tumor diagnosis with improved serum stability and extended imaging window in vivo. Anal. Chim. Acta 2014, 812, 138–144. [Google Scholar] [CrossRef]
- Miller, P.S. Oligonucleoside methylphosphonates as antisense reagents. Nat. Biotechnol. 1991, 9, 358–362. [Google Scholar] [CrossRef]
- Wu, X.; Zhao, Z.; Bai, H.; Fu, T.; Yang, C.; Hu, X.; Liu, Q.; Champanhac, C.; Teng, I.T.; Ye, M.; et al. DNA aptamer selected against pancreatic ductal adenocarcinoma for in vivo imaging and clinical tissue recognition. Theranostics 2015, 5, 985–994. [Google Scholar] [CrossRef] [Green Version]
- Mai, J.; Li, X.; Zhang, G.; Huang, Y.; Xu, R.; Shen, Q.; Lokesh, G.L.; Thiviyanathan, V.; Chen, L.; Liu, H.; et al. DNA thioaptamer with homing specificity to lymphoma bone marrow involvement. Mol. Pharm. 2018, 15, 1814–1825. [Google Scholar] [CrossRef]
- Lee, E.J.; Lim, H.K.; Cho, Y.S.; Hah, S.S. Peptide nucleic acids are an additional class of aptamers. RSC Adv. 2013, 3, 5828–5831. [Google Scholar] [CrossRef]
- Iwamoto, N.; Butler, D.C.D.; Svrzikapa, N.; Mohapatra, S.; Zlatev, I.; Sah, D.W.Y.; Standley, S.M.; Lu, G.; Apponi, L.H.; Kamenetsky, M.F.; et al. Control of phosphorothioate stereochemistry substantially increases the efficacy of antisense oligonucleotides. Nat. Biotechnol. 2017, 35, 845–851. [Google Scholar] [CrossRef]
- Guga, P.; Stec, W.J. Synthesis of phosphorothioate oligonucleotides with stereodefined phosphorothioate linkages. Curr. Protoc. Nucleic Acid Chem. 2003, 14, 4.17.11–4.17.28. [Google Scholar] [CrossRef] [PubMed]
- Knouse, K.W.; deGruyter, J.N.; Schmidt, M.A.; Zheng, B.; Vantourout, J.C.; Kingston, C.; Mercer, S.E.; Mcdonald, I.M.; Olson, R.E.; Zhu, Y.; et al. Unlocking P(V): Reagents for chiral phosphorothioate synthesis. Science 2018, 361, 1234–1238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, D.; Rivas-Bascón, N.; Padial, N.M.; Knouse, K.W.; Zheng, B.; Vantourout, J.C.; Schmidt, M.A.; Eastgate, M.D.; Baran, P.S. Enantiodivergent formation of C–P bonds: Synthesis of P-chiral phosphines and methylphosphonate oligonucleotides. J. Am. Chem. Soc. 2020, 142, 5785–5792. [Google Scholar] [CrossRef] [PubMed]
- Nolte, A.; Klußmann, S.; Bald, R.; Erdmann, V.A.; Fürste, J.P. Mirror-design of L-oligonucleotide ligands binding to L-arginine. Nat. Biotechnol. 1996, 14, 1116–1119. [Google Scholar] [CrossRef]
- Yatime, L.; Maasch, C.; Hoehling, K.; Anderson, G.R.; Vater, A. Structural basis for the targeting of complement anaphylatoxin C5a using a mixed L-RNA/L-DNA aptamer. Nat. Commun. 2015, 6, 6481. [Google Scholar] [CrossRef]
- Sczepanski, J.T.; Joyce, G.F. Binding of a structured D-RNA molecule by and L-RNA aptamer. J. Am. Chem. Soc. 2013, 135, 13290–13293. [Google Scholar] [CrossRef] [Green Version]
- Purschke, W.G.; Hoehlig, K.; Buchner, K.; Zboralski, D.; Schwoebel, F.; Vater, A.; Klussmann, S. Identification and characterization of a mirror-image oligonucleotide that binds and neutralizes sphingosine 1-phosphate, a central mediator of angiogenesis. Biochem. J. 2014, 462, 153–162. [Google Scholar] [CrossRef] [Green Version]
- Vater, A.; Klussmann, S. Turning mirror-image oligonucleotides into drugs: The evolution of Spiegelmer((R)) therapeutics. Drug Discov. Today 2015, 20, 147–155. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Xu, W.; Liu, L.; Zhu, T.F. A synthetic molecular system capable of mirror-image genetic replication and transcription. Nat. Chem. 2016, 8, 698–704. [Google Scholar] [CrossRef]
- Pech, A.; Achenbach, J.; Jahnz, M.; Schülzchen, S.; Jarosch, F.; Bordusa, F.; Klussmann, S. A thermostable D-polymerase for mirror-image PCR. Nucleic Acids Res. 2017, 45, 3997–4005. [Google Scholar] [CrossRef] [Green Version]
- Jiang, W.; Zhang, B.; Fan, C.; Wang, M.; Wang, J.; Deng, Q.; Liu, X.; Chen, J.; Zheng, J.; Liu, L.; et al. Mirror-image polymerase chain reaction. Cell Discov. 2017, 3, 17037. [Google Scholar] [CrossRef]
- Glazier, D.A.; Liao, J.; Roberts, B.L.; Li, X.; Yang, K.; Stevens, C.M.; Tang, W. Chemical synthesis and biological application of modified oligonucleotides. Bioconjug. Chem. 2020, 31, 1213–1233. [Google Scholar] [CrossRef] [PubMed]
- Stein, C.A.; Subasinghe, C.; Shinozuka, K.; Cohen, J.S. Physicochemical properties of phospborothioate oligodeoxynucleotides. Nucleic Acids Res. 1988, 16, 3209–3221. [Google Scholar] [CrossRef] [PubMed]
- Mir, A.; Alterman, J.F.; Hassler, M.R.; Debacker, A.J.; Hudgens, E.; Echeverria, D.; Brodsky, M.H.; Khvorova, A.; Watts, J.K.; Sontheimer, E.J. Heavily and fully modified RNAs guide efficient SpyCas9-mediated genome editing. Nat. Commun. 2018, 9, 2641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Latorre, A.; Latorre, A.; Som, Á. Modified RNAs in CRISPR/Cas9: An old trick works again. Angew. Chem. Int. Ed. Engl. 2016, 55, 3548–3550. [Google Scholar] [CrossRef]
- Hendel, A.; Bak, R.O.; Clark, J.T.; Kennedy, A.B.; Ryan, D.E.; Roy, S.; Steinfeld, I.; Lunstad, B.D.; Kaiser, R.J.; Wilkens, A.B.; et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 2015, 33, 985–989. [Google Scholar] [CrossRef]
- Mishra, S.; Lee, Y.; Park, J.W. Direct quantification of trace amounts of a chronic myeloid leukemia biomarker using locked nucleic acid capture probes. Anal. Chem. 2018, 90, 12824–12831. [Google Scholar] [CrossRef]
- Silvia, F.; Joana, B.; Pedro, M.; Céu, F.; Jesper, W.; Nuno Filipe, A. Mismatch discrimination in fluorescent in situ hybridization using different types of nucleic acids. Appl. Genet. Mol. Biotechnol. 2015, 99, 3961–3969. [Google Scholar] [CrossRef] [Green Version]
- Hardin, J.O.; Milam, V.T. Measuring in situ primary and competitive DNA hybridization activity on microspheres. Biomacromolecules 2013, 14, 986–992. [Google Scholar] [CrossRef] [PubMed]
- Olson, X.; Kotani, S.; Yurke, B.; Graugnard, E.; Hughes, W.L. Kinetics of DNA strand displacement systems with locked nucleic acids. J. Phys. Chem. B 2017, 121, 2594–2602. [Google Scholar] [CrossRef] [PubMed]
- Young, B.E.; Sczepanski, J.T. Heterochiral DNA strand-displacement based on chimeric D/L-oligonucleotides. ACS Synth. Biol. 2019, 8, 2756–2759. [Google Scholar] [CrossRef] [PubMed]
- Crooke, S.T.; Wang, S.; Vickers, T.A.; Shen, W.; Liang, X.-h. Cellular uptake and trafficking of antisense oligonucleotides. Nat. Biotechnol. 2017, 35, 230–237. [Google Scholar] [CrossRef]
- Hyjek-Składanowska, M.; Vickers, T.A.; Napiórkowska, A.; Anderson, B.A.; Tanowitz, M.; Crooke, S.T.; Liang, X.H.; Seth, P.P.; Nowotny, M. Origins of the increased affinity of phosphorothioate-modified therapeutic nucleic acids for proteins. J. Am. Chem. Soc. 2020, 142, 7456–7468. [Google Scholar] [CrossRef]
- Shen, W.; De Hoyos, C.L.; Migawa, M.T.; Vickers, T.A.; Sun, H.; Low, A.; Bell, T.A.; Rahdar, M.; Mukhopadhyay, S.; Hart, C.E.; et al. Chemical modification of PS-ASO therapeutics reduces cellular protein-binding and improves the therapeutic index. Nat. Biotechnol. 2019, 37, 640–650. [Google Scholar] [CrossRef]
- Crooke, S.T.; Vickers, T.A.; Liang, X.H. Phosphorothioate modified oligonucleotide–protein interactions. Nucleic Acids Res. 2020, 48, 5235–5253. [Google Scholar] [CrossRef]
- Juliano, R.L. Intracellular trafficking and endosomal release of oligonucleotides: What we know and what we don’t. Nucleic Acid Ther. 2018, 28, 166–177. [Google Scholar] [CrossRef]
- Vollmer, J.; Jepsen, J.S.; Uhlmann, E.; Schetter, C.; Jurk, M.; Wader, T.; Wullner, M.; Krieg, A.M. Modulation of CpG oligodeoxynucleotide-mediated immune stimulation by locked nucleic acid (LNA). Oligonucleotides 2004, 14, 23–31. [Google Scholar] [CrossRef]
- Fucini, R.V.; Haringsma, H.J.; Deng, P.; Flanagan, W.M.; Willingham, A.T. Adenosine modification may be preferred for reducing siRNA immune stimulation. Nucleic Acid Ther. 2012, 22, 205–210. [Google Scholar] [CrossRef] [Green Version]
- Robbins, M.; Judge, A.; Liang, L.; McClintock, K.; Yaworski, E.; MacLachlan, I. 2′-O-methyl-modified RNAs act as TLR7 antagonists. Mol. Ther. 2007, 15, 1663–1669. [Google Scholar] [CrossRef]
- Ke, Y.; Castro, C.; Choi, J.H. Structural DNA nanotechnology: Artificial nanostructures for biomedical research. Ann. Rev. Biomed. Eng. 2018, 20, 375–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandler, M.; Johnson, M.B.; Panigaj, M.; Afonin, K.A. Innate immune responses triggered by nucleic acids inspire the design of immunomodulatory nucleic acid nanoparticles (NANPs). Curr. Opin. Biotechnol. 2020, 63, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Houlihan, G.; Arangundy-Franklin, S.; Holliger, P. Exploring the chemistry of genetic information storage and propagation through polymerase engineering. Acc. Chem. Res. 2017, 50, 1079–1087. [Google Scholar] [CrossRef] [PubMed]
- Robaldo, L.; Berzal-Herranz, A.; Montserrat, J.M.; Iribarren, A.M. Activity of core-modified 10–23 DNAzymes against HCV. ChemMedChem 2014, 9, 2172–2177. [Google Scholar] [CrossRef]
- Bouchard, P.R.; Hutabarat, R.M.; Thompson, K.M. Discovery and development of therapeutic aptamers. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 237–257. [Google Scholar] [CrossRef]
- Förster, C.; Zydek, M.; Rothkegel, M.; Wu, Z.; Gallin, C.; Geßner, R.; Lisdat, F.; Fürste, J.P. Properties of an LNA-modified ricin RNA aptamer. Biochem. Biophys. Res. Commun. 2012, 419, 60–65. [Google Scholar] [CrossRef]
- Lipi, F.; Chen, S.; Chakravarthy, M.; Rakesh, S.; Veedu, R.N. In vitro evolution of chemically-modified nucleic acid aptamers: Pros and cons, and comprehensive selection strategies. RNA Biol. 2016, 13, 1232–1245. [Google Scholar] [CrossRef] [Green Version]
- Herdewijn, P.; Marlière, P. Toward safe genetically modified organisms through the chemical diversification of nucleic acids. Chem. Biodivers. 2009, 6, 791–808. [Google Scholar] [CrossRef]
- Rangel, A.E.; Chen, Z.; Ayele, T.M.; Heemstra, J.M. In vitro selection of an XNA aptamer capable of small-molecule recognition. Nucleic Acids Res. 2018, 46, 8057–8068. [Google Scholar] [CrossRef]
- Dunn, M.R.; Chaput, J.C. Reverse transcription of threose nucleic acid by a naturally occurring DNA polymerase. ChemBioChem 2016, 17, 1804–1808. [Google Scholar] [CrossRef]
- Wang, Y.; Ngor, A.K.; Nikoomanzar, A.; Chaput, J.C. Evolution of a general RNA-cleaving FANA enzyme. Nat. Commun. 2018, 9, 5067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houlihan, G.; Arangundy-Franklin, S.; Porebski, B.T.; Subramanian, N.; Taylor, A.I.; Holliger, P. Discovery and evolution of RNA and XNA reverse transcriptase function and fidelity. Nat. Chem. 2020, 12, 683–690. [Google Scholar] [CrossRef] [PubMed]
- Potapov, V.; Fu, X.; Dai, N.; Corrêa, I.R., Jr.; Tanner, N.A.; Ong, J.L. Base modifications affecting RNA polymerase and reverse transcriptase fidelity. Nucleic Acids Res. 2018, 46, 5753–5763. [Google Scholar] [CrossRef] [PubMed]
- Gold, L.; Ayers, D.; Bertino, J.; Bock, C.; Bock, A.; Brody, E.N.; Carter, J.; Dalby, A.B.; Eaton, B.E.; Fitzwater, T.; et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE 2010, 5, e15004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gawande, B.N.; Rohloff, J.C.; Carter, J.D.; von Carlowitz, I.; Zhang, C.; Schneider, D.J.; Janjic, N. Selection of DNA aptamers with two modified bases. Proc. Natl. Acad. Sci. USA 2017, 114, 2898–2903. [Google Scholar] [CrossRef] [Green Version]
- Bugaut, A.; Toulme, J.J.; Rayner, B. SELEX and dynamic combinatorial chemistry interplay for the selection of conjugated RNA aptamers. Org. Biomol. Chem. 2006, 4, 4082–4088. [Google Scholar] [CrossRef]
- Vaught, J.D.; Bock, C.; Carter, J.; Fitzwater, T.; Otis, M.; Schneider, D.; Rolando, J.; Waugh, S.; Wilcox, S.K.; Eaton, B.E. Expanding the chemistry of DNA for in vitro selection. J. Am. Chem. Soc. 2010, 132, 4141–4151. [Google Scholar] [CrossRef]
- Tolle, F.; Brandle, G.M.; Matzner, D.; Mayer, G. A versatile approach towards nucleobase-modified aptamers. Angew. Chem. Int. Ed. Engl. 2015, 54, 10971–10974. [Google Scholar] [CrossRef]
- Pfeiffer, F.; Tolle, F.; Rosenthal, M.; Brandle, G.M.; Ewers, J.; Mayer, G. Identification and characterization of nucleobase-modified aptamers by click-SELEX. Nat. Protoc. 2018, 13, 1153–1180. [Google Scholar] [CrossRef]
- Gordon, C.K.L.; Wu, D.; Pusuluri, A.; Feagin, T.A.; Csordas, A.T.; Eisenstein, M.S.; Hawker, C.J.; Niu, J.; Soh, H.T. Click-particle display for base-modified aptamer discovery. ACS Chem. Biol. 2019, 14, 2652–2662. [Google Scholar] [CrossRef] [PubMed]
- Temme, J.S.; MacPherson, I.S.; DeCourcey, J.F.; Krauss, I.J. High temperature SELMA: Evolution of DNA-supported oligomannose clusters which are tightly recognized by HIV bnAb 2G12. J. Am. Chem. Soc. 2014, 136, 1726–1729. [Google Scholar] [CrossRef] [PubMed]
- Temme, J.S.; Krauss, I.J. SELMA: Selection with modified aptamers. Curr. Protoc. Chem. Biol. 2015, 7, 73–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, D.; Movahedi, M.; Mahdavi-Amiri, Y.; Yeung, W.; Tiburcio, T.; Chen, D.; Hili, R. Evolutionary outcomes of diversely functionalized aptamers isolated from in vitro evolution. ACS Synth. Biol. 2020, 9, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Hollenstein, M. Nucleic acid enzymes based on functionalized nucleosides. Curr. Opin. Chem. Biol. 2019, 52, 93–101. [Google Scholar] [CrossRef]
- Sefah, K.; Yang, Z.; Bradley, K.M.; Hoshika, S.; Jimenez, E.; Zhang, L.; Zhu, G.; Shanker, S.; Yu, F.; Turek, D.; et al. In vitro selection with artificial expanded genetic information systems. Proc. Natl. Acad. Sci. USA 2014, 111, 1449–1454. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Yang, Z.; Sefah, K.; Bradley, K.M.; Hoshika, S.; Kim, M.J.; Kim, H.J.; Zhu, G.; Jimenez, E.; Cansiz, S.; et al. Evolution of functional six-nucleotide DNA. J. Am. Chem. Soc. 2015, 137, 6734–6737. [Google Scholar] [CrossRef] [Green Version]
- Kimoto, M.; Yamashige, R.; Matsunaga, K.; Yokoyama, S.; Hirao, I. Generation of high-affinity DNA aptamers using an expanded genetic alphabet. Nat. Biotechnol. 2013, 31, 453–457. [Google Scholar] [CrossRef]
- Matsunaga, K.I.; Kimoto, M.; Hirao, I. High-affinity DNA aptamer generation targeting von Willebrand Factor A1-domain by genetic alphabet expansion for systematic evolution of ligands by exponential enrichment using two types of libraries composed of five different bases. J. Am. Chem. Soc. 2017, 139, 324–334. [Google Scholar] [CrossRef]
- Chen, Z.; Lichtor, P.A.; Berliner, A.P.; Chen, J.C.; Liu, D.R. Evolution of sequence-defined highly functionalized nucleic acid polymers. Nat. Chem. 2018, 10, 420–427. [Google Scholar] [CrossRef]
- Kong, D.; Lei, Y.; Yeung, W.; Hili, R. Enzymatic synthesis of sequence-defined synthetic nucleic acid polymers with diverse functional groups. Angew. Chem. Int. Ed. Engl. 2016, 55, 13164–13168. [Google Scholar] [CrossRef] [Green Version]
- Bock, L.C.; Griffin, L.C.; Latham, J.A.; Vermaas, E.H.; Toole, J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 1992, 355, 564–566. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.; Yeung, W.; Hili, R. In vitro selection of diversely functionalized aptamers. J. Am. Chem. Soc. 2017, 139, 13977–13980. [Google Scholar] [CrossRef] [PubMed]
- Gasse, C.; Zaarour, M.; Noppen, S.; Abramov, M.; Marliere, P.; Liekens, S.; De Strooper, B.; Herdewijn, P. Modulation of BACE1 activity by chemically modified aptamers. ChemBioChem 2018, 19, 754–763. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Zhao, M.; Wang, J.; Li, Z.; Liang, L.; Zhang, L.; Yuan, Q.; Tan, W. Regulation of protein activity and cellular functions mediated by molecularly evolved nucleic acids. Angew. Chem. Int. Ed. Engl. 2019, 58, 1621–1625. [Google Scholar] [CrossRef]
- Santoro, S.W.; Joyce, G.F.; Sakthivel, K.; Gramatikova, S.; Barbas, C.F., III. RNA cleavage by a DNA enzyme with extended chemical functionality. J. Am. Chem. Soc. 2000, 122, 2433–2439. [Google Scholar] [CrossRef]
- Hollenstein, M.; Hipolito, C.J.; Lam, C.H.; Perrin, D.M. A DNAzyme with three protein-like functional groups: Enhancing catalytic efficiency of M2+-independent RNA cleavage. ChemBioChem 2009, 10, 1988–1992. [Google Scholar] [CrossRef]
- Du, S.; Li, Y.; Chai, Z.; Shi, W.; He, J. Site-specific functionalization with amino, guanidinium, and imidazolyl groups enabling the activation of 10–23 DNAzyme. RSC Adv. 2020, 10, 19067–19075. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, E.; Lam, C.H.; Perrin, D.M. A densely modified M2+-independent DNAzyme that cleaves RNA efficiently with multiple catalytic turnover. Chem. Sci. 2018, 9, 1813–1821. [Google Scholar] [CrossRef]
- Zhou, C.; Avins, J.L.; Klauser, P.C.; Brandsen, B.M.; Lee, Y.; Silverman, S.K. DNA-catalyzed amide hydrolysis. J. Am. Chem. Soc. 2016, 138, 2106–2109. [Google Scholar] [CrossRef] [Green Version]
- Taylor, A.I.; Pinheiro, V.B.; Smola, M.J.; Morgunov, A.S.; Peak-Chew, S.; Cozens, C.; Weeks, K.M.; Herdewijn, P.; Holliger, P. Catalysts from synthetic genetic polymers. Nature 2015, 518, 427–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Zhu, L.; Zhu, Z.; Fu, H.; Jenkins, G.; Wang, C.; Zou, Y.; Lu, X.; Yang, C.J. Backbone modification promotes peroxidase activity of G-quadruplex-based DNAzyme. Chem. Commun. 2012, 48, 8347–8349. [Google Scholar] [CrossRef]
- Huang, P.J.; Liu, J. An ultrasensitive light-up Cu2+ biosensor using a new DNAzyme cleaving a phosphorothioate-modified substrate. Anal. Chem. 2016, 88, 3341–3347. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.J.; Liu, J. Rational evolution of Cd2+-specific DNAzymes with phosphorothioate modified cleavage junction and Cd2+ sensing. Nucleic Acids Res. 2015, 43, 6125–6133. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Feng, M.; Xiao, L.; Tong, A.; Xiang, Y. Postsynthetic modification of DNA phosphodiester backbone for photocaged DNAzyme. ACS Chem. Biol. 2016, 11, 444–451. [Google Scholar] [CrossRef]
- Richards, J.L.; Seward, G.K.; Wang, Y.H.; Dmochowski, I.J. Turning the 10-23 DNAzyme on and off with light. ChemBioChem 2010, 11, 320–324. [Google Scholar] [CrossRef] [Green Version]
- Kamiya, Y.; Arimura, Y.; Ooi, H.; Kato, K.; Liang, X.G.; Asanuma, H. Development of visible-light-responsive RNA scissors based on a 10–23 DNAzyme. ChemBioChem 2018, 19, 1305–1311. [Google Scholar] [CrossRef]
- Yang, Z.; Loh, K.Y.; Chu, Y.T.; Feng, R.; Satyavolu, N.S.R.; Xiong, M.; Nakamata Huynh, S.M.; Hwang, K.; Li, L.; Xing, H.; et al. Optical control of metal ion probes in cells and zebrafish using highly selective DNAzymes conjugated to upconversion nanoparticles. J. Am. Chem. Soc. 2018, 140, 17656–17665. [Google Scholar] [CrossRef]
- Chakravarthy, M.; Aung-Htut, M.T.; Le, B.T.; Veedu, R.N. Novel Chemically-modified DNAzyme targeting Integrin alpha-4 RNA transcript as a potential molecule to reduce inflammation in multiple sclerosis. Sci. Rep. 2017, 7, 1613. [Google Scholar] [CrossRef]
- Sczepanski, J.T.; Joyce, G.F. A cross-chiral RNA polymerase ribozyme. Nature 2014, 515, 440–442. [Google Scholar] [CrossRef] [Green Version]
- Afonin, K.A.; Kireeva, M.; Grabow, W.W.; Kashlev, M.; Jaeger, L.; Shapiro, B.A. Co-transcriptional assembly of chemically modified RNA nanoparticles functionalized with siRNAs. Nano Lett. 2012, 12, 5192–5195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.; Lytton-Jean, A.K.; Chen, Y.; Love, K.T.; Park, A.I.; Karagiannis, E.D.; Sehgal, A.; Querbes, W.; Zurenko, C.S.; Jayaraman, M.; et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat. Nanotechnol. 2012, 7, 389–393. [Google Scholar] [CrossRef] [PubMed]
- Jasinski, D.; Haque, F.; Binzel, D.W.; Guo, P. Advancement of the emerging field of RNA nanotechnology. ACS Nano 2017, 11, 1142–1164. [Google Scholar] [CrossRef] [PubMed]
- Tapp, M.J.N.; Slocik, J.M.; Dennis, P.B.; Naik, R.R.; Milam, V.T. Competition-enhanced ligand selection to identify DNA aptamers. ACS Comb. Sci. 2018, 20, 585–593. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, R.; Adams, M.C.; Naik, R.R.; Milam, V.T. Analyzing secondary structure patterns in DNA aptamers identified via CompELS. Molecules 2019, 24, 1572. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Modification | Nuclease Resistance | Polymerase Compatibility | Duplex Stability | Watson–Crick Base Pairing | Ref. | |
|---|---|---|---|---|---|---|
| Sugar | 2′-F | Increase | Yes | Increase | Yes | [17,18] |
| 2′-OMe | Increase | Yes | Increase | Yes | [17,18] | |
| 2′-NH2 | Increase | Yes | Decrease | Yes | [17,18] | |
| LNA | Increase | Yes | Increase | Yes | [18,19] | |
| HNA | Increase | Yes | Increase | Yes | [12] | |
| Phosphodiester Linkage | triazole-linked | Increase | No | Decrease | N/A | [20] |
| PS | Increase | Yes | Decrease | N/A | [21,22] | |
| phNA | Increase | Yes | Decrease | N/A | [23] | |
| Base | 7-deaza-dA | Increase | Yes | Decrease | No | [24] |
| Z/P | Unreported | Yes | Increase | Yes (modified) | [25] | |
| Ds/Px | Unchanged | Yes | Increase | No | [26] | |
| 5-isobutyl-carboxamide-dU | Unreported | Yes | Increase | No | [27] | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ochoa, S.; Milam, V.T. Modified Nucleic Acids: Expanding the Capabilities of Functional Oligonucleotides. Molecules 2020, 25, 4659. https://doi.org/10.3390/molecules25204659
Ochoa S, Milam VT. Modified Nucleic Acids: Expanding the Capabilities of Functional Oligonucleotides. Molecules. 2020; 25(20):4659. https://doi.org/10.3390/molecules25204659
Chicago/Turabian StyleOchoa, Steven, and Valeria T. Milam. 2020. "Modified Nucleic Acids: Expanding the Capabilities of Functional Oligonucleotides" Molecules 25, no. 20: 4659. https://doi.org/10.3390/molecules25204659