Antiproliferation Activity and Mechanism of c9, t11, c15-CLNA and t9, t11, c15-CLNA from Lactobacillus plantarum ZS2058 on Colon Cancer Cells
Abstract
:1. Introduction
2. Results
2.1. Preparation and Purity Detection of CLNA1 and CLNA2
2.2. Anti-proliferative Activity of CLNA1, CLNA2 Against Colon Cancer Cells
2.3. Induction of Lipid Peroxidation by CLNA1 and CLNA2 in Caco-2 Cells
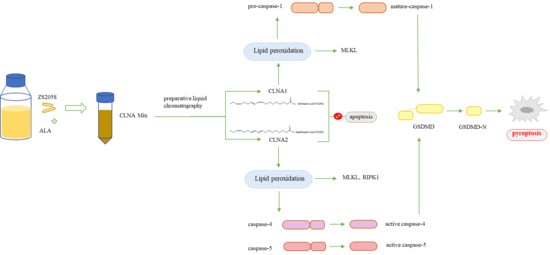
2.4. CLNA1 and CLNA2 Induce Caco-2 Cell Death Independent of Apoptosis and Autophagy
2.5. CLNA1 and CLNA2 Induce Caco-2 Pyroptosis by Different Pathways
3. Discussion
4. Material and Methods
4.1. Microorganism Cultivation and Preparation of CLNA Mixture
4.2. Separation of CLNA and Purity Detection
4.3. Cell Culture
4.4. MTT Cell Viability Assay
4.5. Cell Growth Inhibition Assay
4.6. PI Staining Assay
4.7. Annexin-V/PI Staining Assay
4.8. Western Blotting Analysis
4.9. RNA Extraction and RT-qPCR
4.10. Statistical Analysis
Author Contributions
Funding
Conflicts of Interest
References
- Nagao, K.; Yanagita, T. Conjugated fatty acids in food and their health benefits. J. Biosci. Bioeng. 2005, 100, 152–157. [Google Scholar] [CrossRef]
- Özgül-Yücel, S. Determination of conjugated linolenic acid content of selected oil seeds grown in Turkey. J. Am. Oil Chem. Soc. 2005, 82, 893–897. [Google Scholar] [CrossRef]
- Cao, Y.; Gao, H.L.; Chen, J.N.; Chen, Z.Y.; Yang, L. Identification and Characterization of Conjugated Linolenic Acid Isomers by Ag+-HPLC and NMR. J. Agric. Food Chem. 2006, 54, 9004–9009. [Google Scholar] [CrossRef]
- Shinohara, N.; Tsuduki, T.; Ito, J.; Honma, T.; Kijima, R.; Sugawara, S.; Arai, T.; Yamasaki, M.; Ikezaki, A.; Yokoyama, M.; et al. Jacaric acid, a linolenic acid isomer with a conjugated triene system, has a strong antitumor effect in vitro and in vivo. Biochim. et Biophys. Acta 2012, 1821, 980–988. [Google Scholar] [CrossRef]
- Gasmi, J.; Thomas Sanderson, J. Jacaric acid and its octadecatrienoic acid geoisomers induce apoptosis selectively in cancerous human prostate cells: A mechanistic and 3-D structure-activity study. Phytomedicine 2013, 20, 734–742. [Google Scholar] [CrossRef] [Green Version]
- Paul, D.; Dey, T.K.; Mukherjee, S.; Ghosh, M.; Dhar, P. Comparative prophylactic effects of alpha-eleostearic acid rich nano and conventional emulsions in induced diabetic rats. J. Food Sci. Technol. 2014, 51, 1724–1736. [Google Scholar] [CrossRef] [Green Version]
- Kotwal, S.; Min, J.; Sullivan, D.; Perkovic, V.; Neal, B. Omega 3 Fatty Acids and Cardiovascular Outcomes Systematic Review and Meta-Analysis. Circ. Cardiovasc. Qual. Outcomes 2012, 5, 808–818. [Google Scholar] [CrossRef] [Green Version]
- Vanessa, R.; Jolene, M.M.; Clare, R.; Garry, R.; Martin, R.; Gary, D.; Karen, R.; Graham, H.; Sinead, T.; Moloney, A.P. Inter-organ proteomic analysis reveals insights into the molecular mechanisms underlying the anti-diabetic effects of cis-9, trans-11-conjugated linoleic acid in ob/ob mice. Proteomics 2010, 12, 461. [Google Scholar] [CrossRef]
- Castro-Webb, N.; Ruiz-Narvaez, E.A.; Campos, H. Cross-sectional study of conjugated linoleic acid in adipose tissue and risk of diabetes. Am. J. Clin. Nutr. 2012, 96, 175–181. [Google Scholar] [CrossRef] [Green Version]
- Sengupta, A.; Gupta, S.S.; Nandi, I.; Ghosh, M. Conjugated linolenic acid nanoparticles inhibit hypercholesterolemia induced by feeding a high-fat diet in male albino rats. J. Food Sci. Tech. 2015, 52, 458–464. [Google Scholar] [CrossRef]
- Koba, K.; Imamura, J.; Akashoshi, A.; Kohno-Murase, J.; Nishizono, S.; Iwabuchi, M.; Tanaka, K.; Sugano, M. Genetically Modified Rapeseed Oil Containing cis-9,trans-11,cis-13-Octadecatrienoic Acid Affects Body Fat Mass and Lipid Metabolism in Mice. J. Agric. Food Chem. 2007, 55, 3741. [Google Scholar] [CrossRef]
- Yuan, G.; Chen, X.; Li, D. Modulation of peroxisome proliferator-activated receptor gamma (PPAR gamma) by conjugated fatty acid in obesity and inflammatory bowel disease. J. Agric. Food Chem. 2015, 63, 1883–1895. [Google Scholar] [CrossRef]
- Degen, C.; Ecker, J.; Piegholdt, S.; Liebisch, G.; Schmitz, G.; Jahreis, G. Metabolic and growth inhibitory effects of conjugated fatty acids in the cell line HT-29 with special regard to the conversion of t11,t13-CLA. Biochim. Et Biophys. Acta 2011, 1811, 1070–1080. [Google Scholar] [CrossRef]
- Yasui, Y.; Hosokawa, M.; Kohno, H.; Tanaka, T.; Miyashita, K. Growth inhibition and apoptosis induction by all-trans-conjugated linolenic acids on human colon cancer cells. Anticancer Res. 2006, 26, 1855–1860. [Google Scholar]
- Suzuki, R.; Noguchi, R.; Ota, T.; Abe, M.; Miyashita, K.; Kawada, T. Cytotoxic effect of conjugated trienoic fatty acids on mouse tumor and human monocytic leukemia cells. Lipids 2001, 36, 477–482. [Google Scholar] [CrossRef]
- Grossmann, M.E.; Mizuno, N.K.; Schuster, T.; Cleary, M.P. Punicic acid is an omega-5 fatty acid capable of inhibiting breast cancer proliferation. Int. J. Oncol. 2010, 36, 421–426. [Google Scholar] [CrossRef]
- Grossmann, M.E.; Mizuno, N.K.; Dammen, M.L.; Schuster, T.; Ray, A.; Cleary, M.P. Eleostearic Acid inhibits breast cancer proliferation by means of an oxidation-dependent mechanism. Cancer Prev. Res. 2009, 2, 879. [Google Scholar] [CrossRef] [Green Version]
- Coakley, M.; Banni, S.; Johnson, M.C.; Mills, S.; Devery, R.; Fitzgerald, G.; Paul Ross, R.; Stanton, C.J.L. Inhibitory Effect of Conjugated α-Linolenic Acid from Bifidobacteria of Intestinal Origin on SW480 Cancer Cells. Lipids 2009, 44, 249–256. [Google Scholar] [CrossRef]
- Hennessy, A.A.; Ross, P.; Devery, R.; Stanton, C. Bifidobacterially produced, C18:3 and C18:4 conjugated fatty acids exhibit in vitro anti-carcinogenic and anti-microbial activity. Eur. J. Lipid Sci. Tech. 2016, 118, 1743–1758. [Google Scholar] [CrossRef]
- Yang, B.; Chen, H.; Gao, H.; Ren, Q.; Zhang, H.; Chen, W. Genetic determinates for conjugated linolenic acid production in Lactobacillus plantarum ZS2058. J. Appl. Microbiol. 2019. [Google Scholar] [CrossRef] [Green Version]
- Bailey, K.L.; Agarwal, E.; Chowdhury, S.; Luo, J.; Brattain, M.G.; Black, J.D.; Wang, J. TGFbeta/Smad3 regulates proliferation and apoptosis through IRS-1 inhibition in colon cancer cells. PLoS ONE 2017, 12, e0176096. [Google Scholar] [CrossRef]
- Yasui, Y.; Hosokawa, M.; Sahara, T.; Suzuki, R.; Ohgiya, S.; Kohno, H.; Tanaka, T.; Miyashita, K. Bitter gourd seed fatty acid rich in 9c, 11t, 13t-conjugated linolenic acid induces apoptosis and up-regulates the GADD45, P53 and PPARγ in human colon cancer Caco-2 cells. Prostaglandins Leukot. Essent. Fat. Acids 2005, 73, 113–119. [Google Scholar] [CrossRef]
- Zhai, Q.; Tian, F.; Zhao, J.; Zhang, H.; Narbad, A.; Chen, W. Oral Administration of Probiotics Inhibits Absorption of the Heavy Metal Cadmium by Protecting the Intestinal Barrier. Appl. Environ. Microbiol. 2016, 82, 4429–4440. [Google Scholar] [CrossRef] [Green Version]
- Majid, M.; Nasir, B.; Zahra, S.S.; Khan, M.R.; Mirza, B.; Haq, I.U. Ipomoea batatas L. Lam. ameliorates acute and chronic inflammations by suppressing inflammatory mediators, a comprehensive exploration using in vitro and in vivo models. BMC Complementary Altern. Med. 2018, 18, 216. [Google Scholar] [CrossRef]
- Liu, W.N.; Leung, K.N. Jacaric acid inhibits the growth of murine macrophage-like leukemia PU5-1.8 cells by inducing cell cycle arrest and apoptosis. Cancer Cell Int. 2015, 15, 90. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Gao, W.; Shi, X.; Ding, J.; Liu, W.; He, H.; Wang, K.; Shao, F. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature 2017, 547, 99–103. [Google Scholar] [CrossRef]
- Ding, J.; Shao, F. SnapShot: The noncanonical inflammasome. Cell 2017, 168, 544–544.e1. [Google Scholar] [CrossRef]
- Shi, J.; Zhao, Y.; Wang, K.; Shi, X.; Wang, Y.; Huang, H.; Zhuang, Y.; Cai, T.; Wang, F.; Shao, F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 2015, 526, 660. [Google Scholar] [CrossRef]
- Kayagaki, N.; Stowe, I.B.; Lee, B.L.; O’Rourke, K.; Anderson, K.; Warming, S.; Cuellar, T.; Haley, B.; Roose-Girma, M.; Phung, Q.T. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 2015, 526, 666. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Z.; Ruan, J.; Pan, Y.; Magupalli, V.G.; Wu, H.; Lieberman, J. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 2016, 535, 153. [Google Scholar] [CrossRef] [Green Version]
- Pizato, N.; Luzete, B.C.; Kiffer, L.; Correa, L.H.; de Oliveira Santos, I.; Assumpcao, J.A.F.; Ito, M.K.; Magalhaes, K.G. Omega-3 docosahexaenoic acid induces pyroptosis cell death in triple-negative breast cancer cells. Sci. Rep. 2018, 8, 1952. [Google Scholar] [CrossRef] [PubMed]
- Srikanth, M.; Chandrasaharan, K.; Zhao, X.; Chayaburakul, K.; Ong, W.Y.; Herr, D.R. Metabolism of Docosahexaenoic Acid (DHA) Induces Pyroptosis in BV-2 Microglial Cells. Neuromolecular Med. 2018, 20, 504–514. [Google Scholar] [CrossRef] [PubMed]
- Tsikas, D. Assessment of lipid peroxidation by measuring malondialdehyde (MDA) and relatives in biological samples: Analytical and biological challenges. Anal. Biochem. 2017, 524, 13–30. [Google Scholar] [CrossRef]
- Kang, R.; Zeng, L.; Zhu, S.; Xie, Y.; Liu, J.; Wen, Q.; Cao, L.; Xie, M.; Ran, Q.; Kroemer, G.; et al. Lipid Peroxidation Drives Gasdermin D-Mediated Pyroptosis in Lethal Polymicrobial Sepsis. Cell Host Microbe 2018, 24, 97–108. [Google Scholar] [CrossRef] [Green Version]
- Russo, A.J.; Rathinam, V.A.K. Lipid Peroxidation Adds Fuel to Pyr(optosis). Cell Host Microbe 2018, 24, 8–9. [Google Scholar] [CrossRef] [Green Version]
- Gasmi, J.; Sanderson, J.T. Growth inhibitory, antiandrogenic, and pro-apoptotic effects of punicic acid in LNCaP human prostate cancer cells. J. Agric. Food Chem. 2010, 58, 12149–12156. [Google Scholar] [CrossRef]
- Rathkey, J.K.; Zhao, J.; Liu, Z.; Chen, Y.; Yang, J.; Kondolf, H.C.; Benson, B.L.; Chirieleison, S.M.; Huang, A.Y.; Dubyak, G.R. Chemical disruption of the pyroptotic pore-forming protein gasdermin D inhibits inflammatory cell death and sepsis. Sci. Immunol. 2018, 3, eaat2738. [Google Scholar] [CrossRef] [Green Version]
- Yang, B.; Chen, H.; Stanton, C.; Chen, Y.Q.; Zhang, H.; Chen, W. Mining bifidobacteria from the neonatal gastrointestinal tract for conjugated linolenic acid production. Bioengineered 2017, 8, 232–238. [Google Scholar] [CrossRef] [Green Version]
- Lin, G.; Zhu, S.; Wu, Y.; Song, C.; Wang, W.; Zhang, Y.; Chen, Y.L.; He, Z. omega-3 free fatty acids and all-trans retinoic acid synergistically induce growth inhibition of three subtypes of breast cancer cell lines. Sci. Rep. 2017, 7, 2929. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Chen, H.; Yang, B.; Gu, Z.; Zhang, H.; Chen, W.; Chen, Y.Q. Lactobacillus plantarum ZS2058 produces CLA to ameliorate DSS-induced acute colitis in mice. RSC Adv. 2016, 6, 14457–14464. [Google Scholar] [CrossRef]
Sample Availability: Not available. |












| Cell | ALA | CLNA1 | CLNA2 |
|---|---|---|---|
| Caco-2 | 38.93 ± 2.18 | 18.26 ± 2.06 | 19.75 ± 1.83 |
| SW480 | 21.08 ± 0.67 | 59.46 ± 4.67 | 43.81 ± 11.81 |
| HT-29 | 34.46 ± 1.31 | 47.07 ± 8.22 | 67.52 ± 2.87 |
| Primers | Sequence (5′–3′) |
|---|---|
| GAPDH-F | CCTGGCCAAGGTCATCCATG |
| GAPDH-R | GGAAGGCCATGCCATGGAGC |
| PPARγ-F | ATGGAGCCCAAGTTTGAGTTT |
| PPARγ-R | TGTCTGAGGTCCGTCATTTTC |
| TNF-α-F | ATGAGCACAGAAAGCATGATC |
| TNF-α-R | TACAGGCTTGTCACTCGAATT |
| TGF-β-F | GGCCAGATCCTGTCCAAGC |
| TGF-β-R | GTGGGTTTCCACCATTAGCAC |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, Q.; Yang, B.; Zhu, G.; Wang, S.; Fu, C.; Zhang, H.; Ross, R.P.; Stanton, C.; Chen, H.; Chen, W. Antiproliferation Activity and Mechanism of c9, t11, c15-CLNA and t9, t11, c15-CLNA from Lactobacillus plantarum ZS2058 on Colon Cancer Cells. Molecules 2020, 25, 1225. https://doi.org/10.3390/molecules25051225
Ren Q, Yang B, Zhu G, Wang S, Fu C, Zhang H, Ross RP, Stanton C, Chen H, Chen W. Antiproliferation Activity and Mechanism of c9, t11, c15-CLNA and t9, t11, c15-CLNA from Lactobacillus plantarum ZS2058 on Colon Cancer Cells. Molecules. 2020; 25(5):1225. https://doi.org/10.3390/molecules25051225
Chicago/Turabian StyleRen, Qing, Bo Yang, Guangzhen Zhu, Shunyu Wang, Chengli Fu, Hao Zhang, R. Paul Ross, Catherine Stanton, Haiqin Chen, and Wei Chen. 2020. "Antiproliferation Activity and Mechanism of c9, t11, c15-CLNA and t9, t11, c15-CLNA from Lactobacillus plantarum ZS2058 on Colon Cancer Cells" Molecules 25, no. 5: 1225. https://doi.org/10.3390/molecules25051225