Analysis of Phospholipids, Lysophospholipids, and Their Linked Fatty Acyl Chains in Yellow Lupin Seeds (Lupinus luteus L.) by Liquid Chromatography and Tandem Mass Spectrometry
Abstract
:1. Introduction
2. Results and Discussion
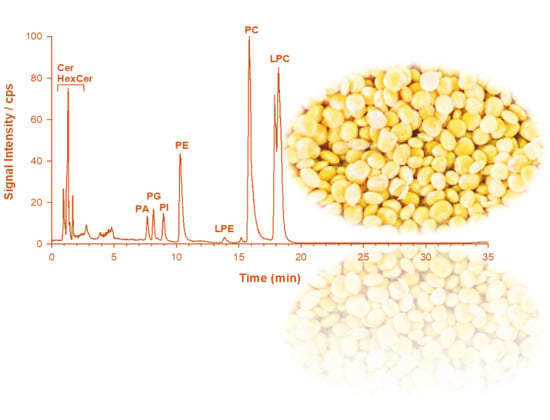
2.1. Phospholipid Analysis of L. luteus Seed Extracts by LC-ESI-FTMS
2.2. CID-MS/MS of Phospholipids in the Lipid Extract of L. luteus Seeds
2.3. Characterization of Lysophospholipids by LC-ESI-MS/MS
2.4. RPLC-ESI-MS/MS of Linked Fatty Acyl Chains upon Chemical Hydrolysis
2.5. Lupin Seeds as an Alternative Source of PL
3. Materials and Methods
3.1. Chemicals
3.2. Sample Preparation
3.2.1. Lipid Extraction
3.2.2. SPE Purification
3.2.3. Chemical Hydrolysis of Linked Fatty Acyl Chains
3.3. LC-ESI-MS Instrumentation and Operating Conditions
3.4. Preliminary Identification of PL by an Online Lipid Calculator
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lupinus Luteus, L. The Plant List. Available online: http://www.theplantlist.org/tpl1.1/record/ild-8560 (accessed on 21 December 2018).
- Finch, H.J.S.; Samuel, A.M.; Lane, G.P.F. Combinable break crops. In Lockhart and Wiseman’s Crop Husbandry Including Grassland; Woodhead Publishing: Cambridge, UK, 2014; pp. 337–361. [Google Scholar]
- Trugo, L.C.; von Baer, E.; von Baer, D. Lupin Breeding. In Reference Module in Food Science; Elsevier: Amsterdam, The Netherlands, 2016; ISBN 9780081005965. [Google Scholar]
- Tapadia, M.; Carlessi, R.; Johnson, S.; Utikar, R.; Newsholme, P. Lupin seed hydrolysate promotes G-protein-coupled receptor, intracellular Ca2+ and enhanced glycolytic metabolism-mediated insulin secretion from BRIN-BD11 pancreatic beta cells. Mol. Cell. Endocrinol. 2019, 480, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Van de Noort, M. Lupin: An Important Protein and Nutrient Source. In Sustainable Protein Sources; Academic Press: Cambridge, MA, USA, 2017; pp. 165–183. [Google Scholar]
- Babini, E.; Tagliazucchi, D.; Martini, S.; Dei Più, L.; Gianotti, A. LC-ESI-QTOF-MS identification of novel antioxidant peptides obtained by enzymatic and microbial hydrolysis of vegetable proteins. Food Chem. 2017, 228, 186–196. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.; Frias, J.; Vidal-Valverde, C. Changes in chemical composition of lupin seeds (Lupinus angustifolius) after selective α-galactoside extraction. J. Sci. Food Agric. 2005, 85, 2468–2474. [Google Scholar] [CrossRef]
- Martínez-Villaluenga, C.; Frías, J.; Vidal-Valverde, C. Functional lupin seeds (Lupinus albus L. and Lupinus luteus L.) after extraction of α-galactosides. Food Chem. 2006, 98, 291–299. [Google Scholar] [CrossRef]
- Eyup Bagci, B.; Bruehl, L.; Özçelik, H.; Aitzetmuller, K.; Vural, M.; Sahim, A. A study of the fatty acid and tocochromanol patterns of some Fabaceae (Leguminosae) plants from Turkey I. Grasas Aceites 2004, 55, 378–384. [Google Scholar]
- Bhardwaj, H.L.; Hamama, A.A.; Van Santen, E. Fatty acids and oil content in white lupin seed as affected by production practices. JAOCS J. Am. Oil Chem. Soc. 2004, 81, 1035–1038. [Google Scholar] [CrossRef]
- Küllenberg, D.; Taylor, L.A.; Schneider, M.; Massing, U. Health Effects of Dietary Phospholipids. Lipids Health Dis. 2012, 11, 3. [Google Scholar] [CrossRef] [Green Version]
- Borek, S.; Pukacka, S.; Michalski, K.; Ratajczak, L. Lipid and protein accumulation in developing seeds of three lupine species: Lupinus luteus L., Lupinus albus L., and Lupinus mutabilis Sweet. J. Exp. Bot. 2009, 60, 3453–3466. [Google Scholar] [CrossRef] [Green Version]
- Hamama, A.A.; Bhardwaj, H.L. Phytosterols, triterpene alcohols, and phospholipids in seed oil from white lupin. JAOCS J. Am. Oil Chem. Soc. 2004, 81, 1039–1044. [Google Scholar] [CrossRef]
- Cajka, T.; Fiehn, O. Comprehensive analysis of lipids in biological systems by liquid chromatography-mass spectrometry. TrAC Trends Anal. Chem. 2014, 61, 192–206. [Google Scholar] [CrossRef] [Green Version]
- Facchini, L.; Losito, I.; Cataldi, T.R.I.; Palmisano, F. Ceramide lipids in alive and thermally stressed mussels: An investigation by hydrophilic interaction liquid chromatography-electrospray ionization Fourier transform mass spectrometry. J. Mass Spectrom. 2016, 51, 768–781. [Google Scholar] [CrossRef] [PubMed]
- Facchini, L.; Losito, I.; Cianci, C.; Cataldi, T.R.I.; Palmisano, F. Structural characterization and profiling of lyso-phospholipids in fresh and in thermally stressed mussels by hydrophilic interaction liquid chromatography-electrospray ionization-Fourier transform mass spectrometry. Electrophoresis 2016, 37, 1823–1838. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.H.; Zou, X.; Huang, J.; Abed, S.M.; Tao, G.; Jin, Q.; Wang, X. Profiling of phospholipids molecular species from different mammalian milk powders by using ultra-performance liquid chromatography-electrospray ionization-quadrupole-time of flight-mass spectrometry. J. Food Compos. Anal. 2017, 62, 143–154. [Google Scholar] [CrossRef]
- Buszewski, B.; Noga, S. Hydrophilic interaction liquid chromatography (HILIC)—A powerful separation technique. Anal. Bioanal. Chem. 2012, 402, 231–247. [Google Scholar] [CrossRef] [Green Version]
- Ventura, G.; Calvano, C.D.; Losito, I.; Bianco, G.; Pascale, R.; Palmisano, F.; Cataldi, T.R.I. Effect of pH and mobile phase additives on the chromatographic behaviour of an amide-embedded stationary phase: Cyanocobalamin and its diaminemonochloro-platinum(II) conjugate as a case study. J. Sep. Sci. 2019, 42, 1155–1162. [Google Scholar] [CrossRef]
- Losito, I.; Facchini, L.; Valentini, A.; Cataldi, T.R.I.; Palmisano, F. Fatty acidomics: Evaluation of the effects of thermal treatments on commercial mussels through an extended characterization of their free fatty acids by liquid chromatography—Fourier transform mass spectrometry. Food Chem. 2018, 255, 309–322. [Google Scholar] [CrossRef]
- Schwalbe-Herrmann, M.; Willmann, J.; Leibfritz, D. Separation of phospholipid classes by hydrophilic interaction chromatography detected by electrospray ionization mass spectrometry. J. Chromatogr. A 2010, 1217, 5179–5183. [Google Scholar] [CrossRef]
- Granafei, S.; Losito, I.; Palmisano, F.; Cataldi, T.R.I.I. Identification of isobaric lyso-phosphatidylcholines in lipid extracts of gilthead sea bream (Sparus aurata) fillets by hydrophilic interaction liquid chromatography coupled to high-resolution Fourier-transform mass spectrometry. Anal. Bioanal. Chem. 2015, 407, 6391–6404. [Google Scholar] [CrossRef]
- Hsu, F.F. Mass spectrometry-based shotgun lipidomics—A critical review from the technical point of view. Anal. Bioanal. Chem. 2018, 410, 6387–6409. [Google Scholar] [CrossRef]
- Hsu, F.-F.; Turk, J. Charge-remote and charge-driven fragmentation processes in diacyl glycerophosphoethanolamine upon low-energy collisional activation: A mechanistic proposal. J. Am. Soc. Mass Spectrom. 2000, 11, 892–899. [Google Scholar] [CrossRef]
- Hsu, F.-F.; Turk, J. Charge-driven fragmentation processes in diacyl glycerophosphatidic acids upon low-energy collisional activation. A mechanistic proposal. J. Am. Soc. Mass Spectrom. 2000, 11, 797–803. [Google Scholar] [CrossRef] [Green Version]
- Hsu, F.-F.; Turk, J. Electrospray ionization with low-energy collisionally activated dissociation tandem mass spectrometry of glycerophospholipids: Mechanisms of fragmentation and structural characterization. J. Chromatogr. B 2009, 877, 2673–2695. [Google Scholar] [CrossRef] [Green Version]
- Dubots, E.; Botté, C.; Boudière, L.; Yamaryo-Botté, Y.; Jouhet, J.; Maréchal, E.; Block, M.A. Role of phosphatidic acid in plant galactolipid synthesis. Biochimie 2012, 94, 86–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, H.Y.; Xue, H.W. Phosphatidic acid plays key roles regulating plant development and stress responses. J. Integr. Plant Biol. 2018, 60, 851–863. [Google Scholar] [CrossRef] [PubMed]
- Řezanka, T.; Sigler, K. Odd-numbered very-long-chain fatty acids from the microbial, animal and plant kingdoms. Prog. Lipid Res. 2009, 48, 206–238. [Google Scholar] [CrossRef] [PubMed]
- Sato, N.; Hagio, M.; Wada, H.; Tsuzuki, M. Requirement of phosphatidylglycerol for photosynthetic function in thylakoid membranes. Proc. Natl. Acad. Sci. USA 2000, 97, 10655–10660. [Google Scholar] [CrossRef] [Green Version]
- Hsu, F.-F.; Turk, J. Studies on phosphatidylglycerol with triple quadrupole tandem mass spectrometry with electrospray ionization: Fragmentation processes and structural characterization. J. Am. Soc. Mass Spectrom. 2001, 12, 1036–1043. [Google Scholar] [CrossRef] [Green Version]
- Mizoi, J.; Nakamura, M.; Nishida, I. A Study of the Physiological Function of Phosphatidylethanolamine in Arabidopsis. In Advanced Research on Plant Lipids; Springer: Dordrecht, The Netherlands, 2003; pp. 377–380. [Google Scholar]
- Van Nieuwenhuyzen, W.; Tomás, M.C. Update on vegetable lecithin and phospholipid technologies. Eur. J. Lipid Sci. Technol. 2008, 110, 472–486. [Google Scholar] [CrossRef]
- Li, J.; Wang, X.; Zhang, T.; Wang, C.; Huang, Z.; Luo, X.; Deng, Y. A review on phospholipids and their main applications in drug delivery systems. Asian J. Pharm. Sci. 2014, 10, 81–98. [Google Scholar] [CrossRef]
- Murphy, R.C.; Axelsen, P.H. Mass spectrometric analysis of long-chain lipids. Mass Spectrom. Rev. 2011, 30, 579–599. [Google Scholar] [CrossRef] [Green Version]
- Huang, Z.-H.; Gage, D.A.; Sweeley, C.C. Characterization of Diacylglycerylphosphocholine Molecular Species by FAB-CAD-MS/MS: A General Method Not Sensitive to the Nature of the Fatty Acyl Groups. J. Am. Soc. Mass Spectrom. 1992, 3, 71–78. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.Y.; Min, H.K.; Moon, M.H. Simultaneous profiling of lysophospholipids and phospholipids from human plasma by nanoflow liquid chromatography-tandem mass spectrometry. Anal. Bioanal. Chem. 2011, 400, 2953–2961. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Cai, X.; Zhao, L.; Xue, X.; Zou, L.; Zhang, X.; Liang, X. Lysophosphatidylcholine profiling of plasma: Discrimination of isomers and discovery of lung cancer biomarkers. Metabolomics 2010, 6, 478–488. [Google Scholar] [CrossRef]
- Lee, H.Y.; Bahn, S.C.; Shin, J.S.; Hwang, I.; Back, K.; Doelling, J.H.; Beungtae Ryu, S. Multiple forms of secretory phospholipase A2 in plants. Prog. Lipid Res. 2005, 44, 52–67. [Google Scholar] [CrossRef] [PubMed]
- Welti, R.; Li, W.; Li, M.; Sang, Y.; Biesiada, H.; Zhou, H.E.; Rajashekar, C.B.; Williams, T.D.; Wang, X. Profiling membrane lipids in plant stress responses: Role of phospholipase Dα in freezing-induced lipid changes in arabidopsis. J. Biol. Chem. 2002, 277, 31994–32002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.; Suh, S.; Kim, S.; Crain, R.C.; Kwak, J.M.; Nam, H.; Lee, Y. Systemic elevation of phosphatidic acid and lysophospholipid levels in wounded plants. Plant J. 1997, 12, 547–556. [Google Scholar] [CrossRef]
- Cowan, A.K. Phospholipids as plant growth regulators. Plant Growth Regul. 2006, 48, 97–109. [Google Scholar] [CrossRef]
- Wi, S.J.; Seo, S.Y.; Cho, K.; Nam, M.H.; Park, K.Y. Lysophosphatidylcholine enhances susceptibility in signaling pathway against pathogen infection through biphasic production of reactive oxygen species and ethylene in tobacco plants. Phytochemistry 2014, 104, 48–59. [Google Scholar] [CrossRef]
- Kole, C. Wild Crop Relatives: Genomic and Breeding Resources; Kole, C., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; ISBN 978-3-642-14386-1. [Google Scholar]
- Hou, Q.; Ufer, G.; Bartels, D. Lipid signalling in plant responses to abiotic stress. Plant. Cell Environ. 2016, 39, 1029–1048. [Google Scholar] [CrossRef]
- Mariani, M.E.; Fidelio, G.D. Secretory Phospholipases A2 in Plants. Front. Plant Sci. 2019, 10, 861. [Google Scholar] [CrossRef]
- Tarasenko, N.A.; Butina, E.A.; Gerasimenko, O.; Nikonovich, Y.N. Investigation into Chemical Composition of Powdered Lupine Seeds. J. Pharm. Sci. Res. 2017, 9, 1002–1006. [Google Scholar]
- Wendel, A.; by Staff, U. Lecithin. In Kirk-Othmer Encyclopedia of Chemical Technology; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2014; pp. 1–19. [Google Scholar]
- Van Hoogevest, P.; Wendel, A. The use of natural and synthetic phospholipids as pharmaceutical excipients. Eur. J. Lipid Sci. Technol. 2014, 116, 1088–1107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Triebl, A.; Weissengruber, S.; Trötzmüller, M.; Lankmayr, E.; Köfeler, H. Quantitative analysis of N-acylphosphatidylethanolamine molecular species in rat brain using solid-phase extraction combined with reversed-phase chromatography and tandem mass spectrometry. J. Sep. Sci. 2016, 39, 2474–2480. [Google Scholar] [CrossRef]
- Calvano, C.D.; Glaciale, M.; Palmisano, F.; Cataldi, T.R.I. Glycosphingolipidomics of donkey milk by hydrophilic interaction liquid chromatography coupled to ESI and multistage MS. Electrophoresis 2018, 39, 1634–1644. [Google Scholar] [CrossRef]
Sample Availability: Samples of the extracted compounds from lupin seeds are available from the authors. |





| Lipid Species | Accurate m/z Value | Adduct | Empirical Formula (M) | Lipid Species |
|---|---|---|---|---|
| 1 | 671.4672 | [M − H]− | C37H68O8P | PA (16:0/18:2) |
| 2 | 673.4825 | [M − H]− | C37H70O8P | PA (16:0/18:1) |
| 3 | 683.4673 | [M − H]− | C38H68O8P | PA (17:0/18:3) PA (19:3/16:0) |
| 4 | 685.4827 | [M − H]− | C38H70O8P | PA (19:2/16:0) PA (17:0/18:2) |
| 5 | 687.4986 | [M − H]− | C38H72O8P | PA (17:0/18:1) PA (19:1/16:0) |
| 6 | 695.4658 | [M − H]− | C39H68O8P | PA (18:2/18:2) PA (18:1/18:3) PA (18:3/18:1) |
| 7 | 697.4816 | [M − H− | C39H70O8P | PA (18:1/18:2) PA (18:2/18:1) PA (18:0/18:3) PA (20:3/16:0) |
| 8 | 699.4978 | [M − H]− | C39H72O8P | PA (18:0/18:2) PA (18:1/18:1) |
| 9 | 701.5136 | [M − H]− | C39H74O8P | PA (18:1/18:0) PA (16:0/20:1) |
| 10 | 705.4511 | [M − H]− | C40H66O8P | PA (19:3/18:3) |
| 11 | 707.4674 | [M − H]− | C40H68O8P | PA (19:2/18:3) |
| 12 | 709.4823 | [M − H]− | C40H70O8P | PA (19:1/18:3) PA (19:2/18:2) PA (19:3/18:1) |
| 13 | 711.4983 | [M − H]− | C40H72O8P | PA (19:1/18:2) PA (19:2/18:1) PA (19:0/18:3) PA (21:3/16:0) |
| 14 | 713.5132 | [M − H]− | C40H74O8P | PA (19:1/18:1) PA (19:0/18:2) PA (19:2/18:0) PA (21:2/16:0) PA (20:2/17:0) |
| 15 | 723.4969 | [M − H]− | C41H72O8P | PA (20:2/18:2) PA (20:3/18:1) PA (20:1/18:3) |
| 16 | 725.5129 | [M − H]− | C41H74O8P | PA (20:1/18:2) PA (20:2/18:1) |
| 17 | 727.5290 | [M − H]− | C41H76O8P | PA (20:1/18:1) PA (18:2/20:0) |
| 18 | 775.5318 | [M − H]− | C45H76O8P | PA (24:5/18:1) PA (24:6/18:0) |
| 19 | 665.4416 | [M − H]− | C34H66O10P | PG (14:0/14:0) PG (12:0/16:0) |
| 20 | 719.4879 | [M − H]− | C38H72O10P | PG (16:1/16:0) PG (18:1/14:0) |
| 21 | 721.5038 | [M − H]− | C38H74O10P | PG (16:0/16:0) |
| 22 | 731.4884 | [M − H]− | C39H72O10P | PG (18:2/15:0) PG (18:1/15:1) PG (16:0/17:2) PG (16:1/17:1) |
| 23 | 733.5033 | [M − H]− | C39H74O10P | PG (15:0/18:1) PG (17:1-16:0) |
| 24 | 741.4721 | [M − H]− | C40H70O10P | PG (16:2/18:2) PG (16:1/18:3) |
| 25 | 743.4891 | [M − H]− | C40H72O10P | PG (18:3/16:0) PG (18:2/16:1) |
| 26 | 745.5048 | [M − H]− | C40H74O10P | PG (16:0/18:2) |
| 27 | 747.5192 | [M − H]− | C40H76O10P | PG (16:0/18:1) |
| 28 | 757.5022 | [M − H]− | C41H74O10P | PG (17:2/18:1) PG (17:1/18:2) |
| 29 | 759.5171 | [M − H]− | C41H76O10P | PG (18:2/17:0) PG (18:1/17:1) PG (19:2/16:0) |
| 30 | 761.5332 | [M − H]− | C41H78O10P | PG (18:1/17:0) PG (19:1/16:0) |
| 31 | 767.4868 | [M − H]− | C42H72O10P | PG (18:3/18:2) |
| 32 | 769.5028 | [M − H]− | C42H74O10P | PG (18:2/18:2) PG (18:1/18:3) |
| 33 | 771.5188 | [M − H]− | C42H76O10P | PG (18:2/18:1) |
| 34 | 773.5341 | [M − H]− | C42H78O10P | PG (18:1/18:1) PG (16:0/20:2) PG (20:2/16:0) |
| 35 | 775.5488 | [M − H]− | C42H80O10P | PG (18:0/18:1) PG (16:0/20:1) |
| 36 | 777.5655 | [M − H]− | C42H82O10P | PG (18:0/18:0) PG (20:0/16:0) |
| 37 | 831.5037 | [M − H]− | C43H76O13P | PI (16:0/18:3) |
| 38 | 833.5196 | [M − H]− | C43H78O13P | PI (16:0/18:2) |
| 39 | 835.5352 | [M − H]− | C43H80O13P | PI (16:0/18:1) |
| 40 | 855.5022 | [M − H]− | C45H76O13P | PI (18:3/18:2) |
| 41 | 857.5183 | [M − H]− | C45H78O13P | PI (18:2/18:2) PI (18:1/18:3) |
| 42 | 859.5342 | [M − H]− | C45H80O13P | PI (18:1/18:2) |
| 43 | 861.5497 | [M − H]− | C45H82O13P | PI (18:1/18:1) PI (18:0/18:2) |
| 44 | 863.5629 | [M − H]− | C45H84O13P | PI (18:0/18:1) |
| 45 | 634.4502 | [M − H]− | C33H65NO8P | PE (12:0/16:0) PE (10:0/18:0) |
| 46 | 674.4722 | [M − H]− | C36H69NO8P | PE (13:0/18:1) PE (15:1/16:0) |
| 47 | 686.4771 | [M − H]− | C37H69NO8P | PE (14:0/18:2) PE (16:1/16:1) |
| 48 | 688.4927 | [M − H]− | C37H71NO8P | PE (14:0/18:1) PE (16:0/16:1) |
| 49 | 702.5085 | [M − H]− | C38H73NO8P | PE (15:0/18:1) PE (16:0/17:1) |
| 50 | 712.4938 | [M − H]− | C39H71NO8P | PE (16:0/18:3) |
| 51 | 714.5097 | [M − H]− | C39H73NO8P | PE (16:0/18:2) PE (16:1-18:1) |
| 52 | 716.5244 | [M − H]− | C39H75NO8P | PE (16:0/18:1) |
| 53 | 718.5385 | [M − H]− | C39H77NO8P | PE (16:0/18:0) |
| 54 | 738.5097 | [M − H]− | C41H73NO8P | PE (18:2/18:2) PE (18:3/18:1) |
| 55 | 740.5246 | [M − H]− | C41H75NO8P | PE (18:2/18:1) |
| 56 | 742.5395 | [M − H]− | C41H77NO8P | PE (18:1/18:1) |
| 57 | 744.5521 | [M − H]− | C41H79NO8P | PE (18:0/18:1) |
| 58 | 754.5402 | [M − H]− | C42H77NO8P | PE (19:2/18:1) PE (19:1/18:2) PE (17:3/20:0) |
| 59 | 766.5387 | [M − H]− | C43H77NO8P | PE (18:0/20:4) PE (18:1/20:3) |
| 60 | 768.5540 | [M − H]− | C43H79NO8P | PE (20:1/18:2) PE (20:2/18:1) PE (18:0/20:3) PE (20:0/18:3) |
| 61 | 770.5699 | [M − H]− | C43H81NO8P | PE (20:1/18:1) PE (20:0/18:2) |
| 62 | 776.5255 | [M − H]− | C44H75NO8P | PE (21:5/18:1) PE (23:6/16:0) |
| 63 | 778.5388 | [M − H]− | C44H77NO8P | PE (21:5/18:0) PE (21:4/18:1) |
| 64 | 794.5709 | [M − H]− | C45H81NO8P | PE (18:1/22:3) PE (18:0/22:4) PE (18:2/22:2) PE (18:3/22:1) |
| 65 | 796.5862 | [M − H]− | C45H83NO8P | PE (22:1/18:2) PE (22:0/18:3) PE (22:2/18:1) |
| 66 | 452.2793 | [M − H]− | C21H43NO7P | LPE (16:0/0:0) |
| 67 | 452.2793 | [M − H]− | C21H43NO7P | LPE (0:0/16:0) |
| 68 | 474.2639 | [M − H]− | C23H41NO7P | LPE (18:3/0:0) |
| 69 | 474.2639 | [M − H]− | C23H41NO7P | LPE (0:0/18:3) |
| 70 | 476.2792 | [M − H]− | C23H43NO7P | LPE (18:2/0:0) |
| 71 | 476.2792 | [M − H]− | C23H43NO7P | LPE (0:0/18:2) |
| 72 | 478.2947 | [M − H]− | C23H45NO7P | LPE (18:1/0:0) |
| 73 | 478.2947 | [M − H]− | C23H45NO7P | LPE (0:0/18:1) |
| 74 | 480.3055 | [M − H]− | C23H47NO7P | LPE (18:0/0:0) |
| 75 | 480.3055 | [M − H]− | C23H47NO7P | LPE (0:0/18:0) |
| 76 | 506.3262 | [M − H]− | C25H49NO7P | LPE (20:1/0:0) |
| 77 | 506.3262 | [M − H]− | C25H49NO7P | LPE (0:0/20:1) |
| 78 | 536.3734 | [M − H]− | C27H55NO7P | LPE (0:0/22:0) |
| 79 | 712.4932 | [M − CH3]− | C39H71NO8P | PC (14:2/18:1) PC (14:1/18:2) PC (14:0/18:3) |
| 80 | 714.5087 | [M − CH3]− | C39H73NO8P | PC (14:0/18:2) PC (16:1/16:1) |
| 81 | 716.5245 | [M − CH3]− | C39H75NO8P | PC (18:1/14:0) PC (16:0/16:1) |
| 82 | 718.5407 | [M − CH3]− | C39H77NO8P | PC (16:0/16:0) |
| 83 | 740.5252 | [M − CH3]− | C41H75NO8P | PC (16:0/18:3) PC (16:1/18:2) |
| 84 | 742.5414 | [M − CH3]− | C41H77NO8P | PC (16:0/18:2) PC (18:1/16:1) |
| 85 | 744.5568 | [M − CH3]− | C41H79NO8P | PC (16:0/18:1) |
| 86 | 766.5404 | [M − CH3]− | C43H77NO8P | PC (18:2/18:2) PC (18:3-18:1) |
| 87 | 768.5563 | [M − CH3]− | C43H79NO8P | PC (18:2/18:1) |
| 88 | 770.5711 | [M − CH3]− | C43H81NO86P | PC (18:1/18:1) |
| 89 | 778.5607 | [M + HCOO]− | C41H81NO10P | PC (16:0/16:0) |
| 90 | 790.5609 | [M + HCOO]− | C42H81NO10P | PC (15:0/18:1) PC (17:1/16:0) |
| 91 | 792.5766 | [M + HCOO]− | C42H83NO10P | PC (17:0/16:0) |
| 92 | 794.5707 | [M − CH3]− | C45H81NO8P | PC (18:3/20:1) PC (18:2/20:2) PC (18:1/20:3) |
| 93 | 796.5860 | [M − CH3]− | C45H83NO8P | PC (20:1/18:2) PC (20:2/18:1) |
| 94 | 802.5621 | [M + HCOO]− | C43H81NO10P | PC (16:0/18:2) PC (16:1/18:1) |
| 95 | 804.5772 | [M + HCOO]− | C43H83NO10P | PC (16:0/18:1) |
| 96 | 806.5891 | [M + HCOO]− | C43H85NO10P | PC (16:0/18:0) |
| 97 | 826.5620 | [M + HCOO]− | C45H81NO10P | PC (18:2/18:2) PC (18:1/18:3) |
| 98 | 828.5774 | [M + HCOO]− | C45H83NO10P | PC (18:2/18:1) |
| 99 | 830.5918 | [M + HCOO]− | C45H85NO10P | PC (18:1/18:1) |
| 100 | 832.6054 | [M + HCOO]− | C45H87NO10P | PC (18:0/18:1) |
| 101 | 856.6060 | [M + HCOO]− | C47H87NO10P | PC (18:2/20:1) PC (18:1/20:2) |
| 102 | 858.6216 | [M + HCOO]− | C47H89NO10P | PC (20:1/18:1) PC (20:0/18:2) PC (19:1/19:1) |
| 103 | 884.6380 | [M + HCOO]− | C49H91NO10P | PC (18:2/22:1) PC (18:3/22:0) PC (18:1/22:2) |
| 104 | 886.6539 | [M + HCOO]− | C49H93NO10P | PC (18:2/22:0) PC (18:1/22:1) |
| 105 | 888.6692 | [M + HCOO]− | C49H95NO10P | PC (18:1/22:0) |
| 106 | 910.6549 | [M + HCOO]− | C51H93NO10P | PC (24:1/18:3) PC (24:2/18:2) PC (24:3/18:1) |
| 107 | 914.6864 | [M + HCOO]− | C51H97NO10P | PC (24:0/18:2) PC (24:1/18:1) |
| 108 | 916.7018 | [M + HCOO]− | C51H99NO10P | PC (18:1/24:0) |
| 109 | 480.3104 | [M − CH3]− | C23H47NO7P | LPC (16:0/0:0) |
| 110 | 480.3104 | [M − CH3]− | C23H47NO7P | LPC (0:0/16:0) |
| 111 | 504.3109 | [M − CH3]− | C25H47NO7P | LPC (18:2/0:0) |
| 112 | 504.3109 | [M − CH3]− | C25H47NO7P | LPC (0:0/18.2) |
| 113 | 506.3253 | [M − CH3]− | C25H49NO7P | LPC (18:1/0:0) |
| 114 | 506.3253 | [M − CH3]− | C25H49NO7P | LPC (0:0/18:1) |
| 115 | 534.3567 | [M − CH3]− | C27H53NO7P | LPC (20:1/0:0) |
| 116 | 534.3567 | [M − CH3]− | C27H53NO7P | LPC (0:0/20:1) |
| 117 | 536.3725 | [M − CH3]− | C27H55NO7P | LPC (20:0/0:0) |
| 118 | 536.3725 | [M − CH3]− | C27H55NO7P | LPC (0:0/20:0) |
| 119 | 540.3312 | [M + HCOO]− | C25H51NO9P | LPC (16:0/0:0) |
| 120 | 540.3312 | [M + HCOO]− | C25H51NO9P | LPC (0:0/16:0) |
| 121 | 564.3311 | [M + HCOO]− | C27H51NO9P | LPC (18:2/0:0) |
| 122 | 564.3311 | [M + HCOO]− | C27H51NO9P | LPC (0:0/18:2) |
| 123 | 566.3465 | [M + HCOO]− | C27H53NO9P | LPC (18:1/0:0) |
| 124 | 566.3465 | [M + HCOO]− | C27H53NO9P | LPC (0:0/18:1) |
| Accurate m/z | Theoretical m/z | Accuracy (ppm) | Time (min) | FA Composition | Empirical Formula (M) | Relative Abundance (%) |
|---|---|---|---|---|---|---|
| 199.1706 | 199.1704 | +1.0 | 4.55 | 12:0 | C12H23O2 | 0.02 ± 0.01 |
| 227.2018 | 227.2017 | +0.4 | 8.40 | 14:0 | C14H27O2 | 0.15 ± 0.01 |
| 241.2177 | 241.2173 | +1.7 | 11.69 | 15:0 | C15H29O2 | 0.11 ± 0.02 |
| 253.2179 | 253.2173 | +2.4 | 10.16 | 16:1 | C16H29O2 | 0.74 ± 0.03 |
| 255.2332 | 255.2330 | +0.8 | 15.08 | 16:0 | C16H31O2 | 6.84 ± 0.08 |
| 267.2339 | 267.2330 | +3.4 | 13.72 | 17:1 | C17H31O2 | 0.09 ± 0.02 |
| 269.2493 | 269.2486 | +2.6 | 20.15 | 17:0 | C17H33O2 | 0.08 ± 0.01 |
| 271.2284 | 271.2279 | +1.8 | 7.96 | 16:0;1 | C16H31O3 | 0.39 ± 0.13 |
| 277.2173 | 277.2173 | 0.0 | 9.13 | 18:3 | C18H29O2 | 9.36 ± 0.19 |
| 279.2330 | 279.2330 | 0.0 | 12.30 | 18:2 | C18H31O2 | 21.4 ± 1.6 |
| 281.2487 | 281.2486 | +0.4 | 16.96 | 18:1 | C18H33O2 | 38.2 ± 4.5 |
| 283.2642 | 283.2643 | −0.4 | 24.22 | 18:0 | C18H35O2 | 1.23 ± 0.03 |
| 305.2491 | 305.2486 | +1.6 | 15.95 | 20:3 | C20H33O2 | 0.11 ± 0.01 |
| 307.2646 | 307.2643 | +1.0 | 20.58 | 20:2 | C20H35O2 | 0.67 ± 0.12 |
| 309.2801 | 309.2799 | +0.6 | 25.67 | 20:1 | C20H37O2 | 6.03 ± 0.19 |
| 311.2957 | 311.2956 | +0.3 | 33.10 | 20:0 | C20H39O2 | 1.03 ± 0.16 |
| 325.2749 | 325.2748 | +0.3 | 14.99 | 20:1;1 | C20H37O3 | 0.80 ± 0.20 |
| 325.3116 | 325.3112 | +1.2 | 37.23 | 21:0 | C21H41O2 | 0.18 ± 0.05 |
| 333.2802 | 333.2799 | +0.9 | 24.08 | 22:3 | C22H37O2 | 0.28 ± 0.04 |
| 335.2958 | 335.2956 | +0.6 | 28.92 | 22:2 | C22H39O2 | 0.39 ± 0.06 |
| 337.3114 | 337.3112 | +0.6 | 34.00 | 22:1 | C22H41O2 | 3.5 ± 0.5 |
| 339.3266 | 339.3269 | −0.9 | 40.41 | 22:0 | C22H43O2 | 4.0 ± 0.9 |
| 353.3065 | 353.3061 | +1.1 | 21.33 | 22:1;1 | C22H41O3 | 2.6 ± 0.2 |
| 353.3427 | 353.3425 | +0.6 | 44.08 | 23:0 | C23H45O2 | 0.27 ± 0.11 |
| 355.3226 | 355.3218 | +2.3 | 16.32 | 22:0;1 | C22H43O3 | 0.22 ± 0.10 |
| 361.3121 | 361.3112 | +2.5 | 36.30 | 24:3 | C24H41O2 | 0.25 ± 0.04 |
| 363.3275 | 363.3269 | +1.7 | 39.18 | 24:2 | C24H43O2 | 0.06 ± 0.01 |
| 365.3429 | 365.3425 | +1.1 | 41.42 | 24:1 | C24H45O2 | 0.20 ± 0.04 |
| 367.3583 | 367.3582 | +0.3 | 47.00 | 24:0 | C24H47O2 | 1.2 ± 0.5 |
| 383.3539 | 383.3531 | +2.1 | 38.11 | 24:0;1 | C24H47O3 | 0.09 ± 0.05 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calvano, C.D.; Bianco, M.; Ventura, G.; Losito, I.; Palmisano, F.; Cataldi, T.R.I. Analysis of Phospholipids, Lysophospholipids, and Their Linked Fatty Acyl Chains in Yellow Lupin Seeds (Lupinus luteus L.) by Liquid Chromatography and Tandem Mass Spectrometry. Molecules 2020, 25, 805. https://doi.org/10.3390/molecules25040805
Calvano CD, Bianco M, Ventura G, Losito I, Palmisano F, Cataldi TRI. Analysis of Phospholipids, Lysophospholipids, and Their Linked Fatty Acyl Chains in Yellow Lupin Seeds (Lupinus luteus L.) by Liquid Chromatography and Tandem Mass Spectrometry. Molecules. 2020; 25(4):805. https://doi.org/10.3390/molecules25040805
Chicago/Turabian StyleCalvano, Cosima Damiana, Mariachiara Bianco, Giovanni Ventura, Ilario Losito, Francesco Palmisano, and Tommaso R.I. Cataldi. 2020. "Analysis of Phospholipids, Lysophospholipids, and Their Linked Fatty Acyl Chains in Yellow Lupin Seeds (Lupinus luteus L.) by Liquid Chromatography and Tandem Mass Spectrometry" Molecules 25, no. 4: 805. https://doi.org/10.3390/molecules25040805