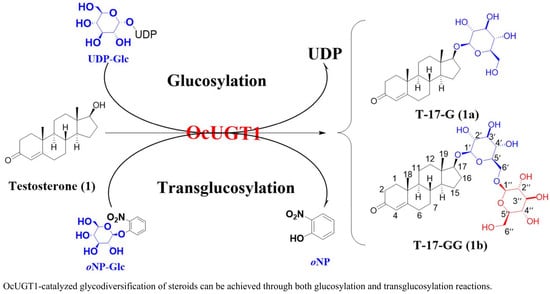
OcUGT1-Catalyzing Glycodiversification of Steroids through Glucosylation and Transglucosylation Actions
Abstract
:1. Introduction
2. Results and Discussion
2.1. Intracellular Expression and Purification of OcUGT1
2.2. OcUGT1-Catalyzed Glucosylation Towards Steroids
2.3. OcUGT1-Mediated Transglucosylation Towards Steroids
3. Materials and Methods
3.1. Plasmids and Strains
3.2. Chemicals and Reagents
3.3. Intracellular Expression and Purification of OcUGT1
3.4. Assays for Glucosylation Activity
3.5. Assays for OcUGT1-Catalyzed Transglucosylation Action
3.6. Analyses and Structural Identification of Steroidal Glucosides
Abbreviations
| DMSO: | Dimethyl sulfoxide |
| GTs: | Glycosyltransferases |
| HPLC: | High-performance liquid chromatography |
| NMR: | Nuclear magnetic resonance |
| oNPGlc: | oortho-nitrophenyl-β-d-glucopyranoside |
| SGs: | Steroidal glycosides |
| SGTs: | Steroidal glycosyltransferases |
| T-17-G: | Testosterone 17-O-β-glucoside |
| T-17-GG: | Testosterone 17-O-β-glucopyranosyl-(1→6)-β-d-glucopyranoside |
| UDP-Glc: | Uridine diphosphate-d-glucose |
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zhang, Y.; Yu, H.-Y.; Chao, L.-P.; Qu, L.; Ruan, J.-Y.; Liu, Y.-X.; Dong, Y.-Z.; Han, L.-F.; Wang, T. Anti-inflammatory steroids from the rhizomes of Dioscorea septemloba Thunb. Steroids 2016, 112, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Xiang, L.; Wang, Y.; Yi, X.; He, X. Anti-inflammatory steroidal glycosides from the berries of Solanum nigrum L. (European black nightshade). Phytochemistry 2018, 148, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.-W.; Zhang, X.; Gong, T.; Gao, W.; Yuan, S.; Zhang, P.-C.; Kong, J.-Q. Structure and bioactivity of cholestane glycosides from the bulbs of Ornithogalum saundersiae Baker. Phytochemistry 2019, 164, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Cimmino, A.; Mathieu, V.; Evidente, M.; Ferderin, M.; Moreno, Y.; Banuls, L.; Masi, M.; De Carvalho, A.; Kiss, R.; Evidente, A. Glanduliferins A and B, two new glucosylated steroids from Impatiens glandulifera, with in vitro growth inhibitory activity in human cancer cells. Fitoterapia 2016, 109, 138–145. [Google Scholar] [CrossRef]
- Peng, Y.-r.; Li, Y.-b.; Liu, X.-d.; Zhang, J.-f.; Duan, J.-a. Antitumor activity of C-21 steroidal glycosides from Cynanchum auriculatum Royle ex Wight. Phytomedicine 2008, 15, 1016–1020. [Google Scholar] [CrossRef]
- Chludil, H.D.; Seldes, A.M.; Maier, M.S. Antifungal steroidal glycosides from the Patagonian Starfish Anasterias minuta: Structure-activity correlations. J. Nat. Prod. 2002, 65, 153–157. [Google Scholar] [CrossRef]
- Favel, A.; Kemertelidze, E.; Benidze, M.; Fallague, K.; Regli, P. Antifungal activity of steroidal glycosides from Yucca gloriosa L. Phytother. Res. 2005, 19, 158–161. [Google Scholar] [CrossRef]
- Singh, O.M.; Subharani, K.; Singh, N.; Devi, N.B.; Nevidita, L. Isolation of steroidal glycosides from Solanum xanthocarpum and studies on their antifungal activities. Nat. Prod. Res. 2007, 21, 585–590. [Google Scholar] [CrossRef]
- Arthan, D.; Svasti, J.; Kittakoop, P.; Pittayakhachonwut, D.; Tanticharoen, M.; Thebtaranonth, Y. Antiviral isoflavonoid sulfate and steroidal glycosides from the fruits of Solanum torvum. Phytochemistry 2002, 59, 459–463. [Google Scholar] [CrossRef]
- Ikeda, T.; Ando, J.; Miyazono, A.; Zhu, X.-H.; Tsumagari, H.; Nohara, T.; Yokomizo, K.; Uyeda, M. Anti-herpes virus activity of Solanum steroidal glycosides. Biol. Pharm. Bull. 2000, 23, 363–364. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Kong, J.-Q. The enzymatic biosynthesis of acylated steroidal glycosides and their cytotoxic activity. Acta Pharm. Sin. B 2018, 8, 981–994. [Google Scholar] [CrossRef] [PubMed]
- Thibodeaux, C.J.; Melancon, C.E.; Liu, H.W. Unusual sugar biosynthesis and natural product glycodiversification. Nature 2007, 446, 1008–1016. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Chen, D.; Chen, R.; Xie, K.; Liu, J.; Yang, L.; Dai, J. Exploring the aglycon promiscuity of a new glycosyltransferase from Pueraria lobata. Tetrahedron Lett. 2016, 57, 1518–1521. [Google Scholar] [CrossRef]
- Chen, D.; Chen, R.; Wang, R.; Li, J.; Xie, K.; Bian, C.; Sun, L.; Zhang, X.; Liu, J.; Yang, L.; et al. Probing the Catalytic Promiscuity of a Regio- and Stereospecific C-Glycosyltransferase from Mangifera indica. Angew. Chem. Int. Ed. Engl. 2015, 54, 12678–12682. [Google Scholar] [CrossRef]
- Xie, K.; Chen, R.; Li, J.; Wang, R.; Chen, D.; Dou, X.; Dai, J. Exploring the catalytic promiscuity of a new glycosyltransferase from Carthamus tinctorius. Org. Lett. 2014, 16, 4874–4877. [Google Scholar] [CrossRef]
- Pandey, R.P.; Bashyal, P.; Parajuli, P.; Yamaguchi, T.; Sohng, J.K. Two trifunctional leloir glycosyltransferases as biocatalysts for natural products glycodiversification. Org. Lett. 2019, 21, 8058–8064. [Google Scholar] [CrossRef]
- Zhu, X.-L.; Wen, C.; Ye, Q.-M.; Xu, W.; Zou, D.-L.; Liang, G.-P.; Zhang, F.; Chen, W.-N.; Jiang, R.-W. Probing the stereoselectivity of OleD-catalyzed glycosylation of cardiotonic steroids. RSC Adv. 2018, 8, 5071–5078. [Google Scholar] [CrossRef] [Green Version]
- Wen, C.; Huang, W.; Zhu, X.-L.; Li, X.-S.; Zhang, F.; Jiang, R.-W. UGT74AN1, a permissive glycosyltransferase from Asclepias curassavica for the regiospecific steroid 3-O-glycosylation. Org. Lett. 2018, 20, 534–537. [Google Scholar] [CrossRef]
- Singh, G.; Dhar, Y.V.; Asif, M.H.; Misra, P. Exploring the functional significance of sterol glycosyltransferase enzymes. Prog. Lipid Res. 2018, 69, 1–10. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, Y.; Feng, Y. Structural dissection of sterol glycosyltransferase UGT51 from Saccharomyces cerevisiae for substrate specificity. J. Struct. Biol. 2018, 204, 371–379. [Google Scholar] [CrossRef]
- Li, K.; Feng, J.; Kuang, Y.; Song, W.; Zhang, M.; Ji, S.; Qiao, X.; Ye, M. Enzymatic synthesis of bufadienolide O-glycosides as potent antitumor agents using a microbial glycosyltransferase. Adv. Synth. Catal. 2017, 359, 3765–3772. [Google Scholar] [CrossRef]
- Hoang, N.H.; Hong, S.Y.; Huong, N.L.; Park, J.W. Biochemical characterization of recombinant UDP-glucose:sterol 3-O-glycosyltransferase from Micromonospora rhodorangea ATCC 31603 and enzymatic biosynthesis of sterol-3-O-beta-glucosides. J. Microbiol. Biotechnol. 2016, 26, 477–482. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Yin, S.; Liu, M.; Kong, J.-Q. Isolation and characterization of a multifunctional flavonoid glycosyltransferase from Ornithogalum caudatum with glycosidase activity. Sci. Rep. 2018, 8, 5886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, S.; Xu, Y.-L.; Yang, Y.; Kong, J.-Q. OcUGT1-catalyzed glucosylation of sulfuretin yields ten glucosides. Catalysts 2018, 8, 416. [Google Scholar] [CrossRef] [Green Version]
- Yuan, S.; Yin, S.; Liu, M.; He, J.-M.; Kong, J.-Q. OcUGT1-catalyzed glycosylation of testosterone with alternative donor substrates. Process Biochem. 2018, 73, 82–85. [Google Scholar] [CrossRef]
- Yuan, S.; Yang, Y.; Kong, J.-Q. Biosynthesis of 7, 8-dihydroxyflavone glycosides via OcUGT1-catalyzed glycosylation and transglycosylation. J. Asian Nat. Prod. Res. 2018, 20, 662–674. [Google Scholar] [CrossRef]
- Liu, M.; Li, L.-N.; Pan, Y.-T.; Kong, J.-Q. cDNA isolation and functional characterization of squalene synthase gene from Ornithogalum caudatum. Protein Expres. Purif. 2017, 130, 63–72. [Google Scholar] [CrossRef]
- Liu, X.; Kong, J.-q. Steroids hydroxylation catalyzed by the monooxygenase mutant 139-3 from Bacillus megaterium BM3. Acta Pharm. Sin. B 2017, 7, 510–516. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds used in this study are available from the authors. |







| Compound 1a | ||
|---|---|---|
| Position | δC | δH |
| 1 | 36.7 | 1.72 (1H, ddd, J = 14.4, 12.6, 4.2 Hz, H-1α) |
| 2.10 (1H, ddd, J = 12.6, 4.8, 3.0 Hz, H-1β) | ||
| 2 | 34.7 | 2.27(1H, ddd, J = 16.2, 4.2, 3.0 Hz, H-2α) |
| 2.46 (1H, ddd, J = 16.2, 14.4, 4.8 Hz, H-2β) | ||
| 3 | 202.3 | - |
| 4 | 124.1 | 5.70 (1H, s, H-4) |
| 5 | 175.2 | - |
| 6 | 33.9 | 2.32 (1H, ddd, J = 14.4, 4.2, 2.4 Hz, H-6α) |
| 2.50 (1H, ddd, J = 14.4, 13.8, 5.4 Hz, H-6β) | ||
| 7 | 32.8 | 1.04 (1H, dddd, J = 13.8, 12.6, 11.4, 4.2 Hz, H-7α) |
| 2.02 (1H, dddd, J = 12.6, 5.4, 3.0, 2.4 Hz, H-7β) | ||
| 8 | 36.8 | 1.68 (1H, dddd, J = 11.4, 10.8, 10.2, 3.0 Hz, H-8) |
| 9 | 55.5 | 0.97 (1H, ddd, J = 12.0, 10.2, 4.2 Hz, H-9) |
| 10 | 40.0 | - |
| 11 | 21.8 | 1.62 (1H, dddd, J = 13.2, 4.2, 4.2, 3.0 Hz, H-11α) |
| 1.50 (1H, dddd, J = 13.8, 13.2, 12.0, 4.2 Hz, H-11β) | ||
| 12 | 38.5 | 1.21 (1H, ddd, J = 13.8, 12.3, 4.2 Hz, H-12α) |
| 1.89 (1H, ddd, J = 12.3, 4.2, 3.0 Hz, H-12β) | ||
| 13 | 44.2 | - |
| 14 | 51.7 | 1.01 (1H, ddd, J = 12.3, 10.8, 7.2 Hz, H-14) |
| 15 | 24.2 | 1.64 (1H, dddd, J = 12.6, 9.6, 7.2, 3.6 Hz, H-15α) |
| 1.32 (1H, dddd, J = 12.6, 12.3, 12.0, 6.0 Hz, H-15β) | ||
| 16 | 29.8 | 1.60 (1H, dddd, J = 13.8, 12.0, 8.4, 3.6 Hz, H-16α) |
| 2.06 (1H, dddd, J = 13.8, 9.6, 9.0, 6.0 Hz, H-16β) | ||
| 17 | 89.6 | 3.76 (1H, dd, J = 9.0, 8.4 Hz, H-17) |
| 18 | 12.0 | 0.90 (3H, s, H-18) |
| 19 | 17.7 | 1.24 (3H, s, H-19) |
| 1ʹ | 104.7 | 4.32 (1H, d, J = 7.8 Hz, H-1ʹ) |
| 2ʹ | 75.4 | 3.15 (1H, dd, J = 9.0, 7.8 Hz, H-2ʹ) |
| 3ʹ | 77.9 | 3.21 (1H, t, J = 9.0 Hz, H-3ʹ) |
| 4ʹ | 71.7 | 3.27 (1H, dd, J = 9.0, 8.4 Hz, H-4ʹ) |
| 5ʹ | 78.2 | 3.33 (1H, dd, J = 8.4, 5.4 Hz, H-5ʹ) |
| 6ʹ | 62.8 | 3.86 (1H, dd, J = 12.0, 2.4 Hz, H-6ʹa) |
| 3.65 (1H, dd, J = 12.0, 5.4 Hz, H-6ʹb) | ||
| Compound 1b | ||
|---|---|---|
| Position | δC | δH |
| 1 | 36.8 | 1.75 (1H, ddd, J = 14.4, 12.6, 4.2 Hz, H-1α) |
| 2.12 (1H, ddd, J = 12.6, 4.8, 3.0 Hz, H-1β) | ||
| 2 | 34.8 | 2.27 (1H, ddd, J = 16.2, 4.2, 3.0 Hz, H-2α) |
| 2.50 (1H, ddd, J = 16.2, 14.4, 4.8 Hz, H-2β) | ||
| 3 | 202.4 | - |
| 4 | 124.1 | 5.70 (1H, s, H-4) |
| 5 | 175.3 | - |
| 6 | 33.9 | 2.33 (1H, ddd, J = 14.2, 4.2, 2.4 Hz, H-6α) |
| 2.50 (1H, ddd, J = 14.2, 13.8, 5.4 Hz, H-6β) | ||
| 7 | 32.8 | 1.08 (1H, dddd, J = 13.8, 12.6, 11.4, 4.2 Hz, H-7α) |
| 2.04 (1H, dddd, J = 12.6, 5.4, 3.0, 2.4 Hz, H-7β) | ||
| 8 | 36.8 | 1.70 (1H, dddd, J = 11.4, 10.8, 10.2, 3.0 Hz, H-8) |
| 9 | 55.4 | 0.99 (1H, ddd, J = 12.0, 10.2, 4.2 Hz, H-9) |
| 10 | 40.0 | - |
| 11 | 21.8 | 1.64 (1H, dddd, J = 13.2, 4.2, 4.2, 3.0 Hz, H-11α) |
| 1.53 (1H, dddd, J = 13.8, 13.2, 12.0, 4.2 Hz, H-11β) | ||
| 12 | 38.3 | 1.23 (1H, ddd, J = 13.8, 12.3, 4.2 Hz, H-12α) |
| 1.91 (1H, ddd, J = 12.3, 4.2, 3.0 Hz, H-12β) | ||
| 13 | 44.2 | - |
| 14 | 51.6 | 1.04 (1H, ddd, J = 12.3, 10.8, 7.2 Hz, H-14) |
| 15 | 24.2 | 1.67 (1H, dddd, J = 12.6, 9.6, 7.2, 3.6 Hz, H-15α) |
| 1.34 (1H, dddd, J = 12.6, 12.3, 12.0, 6.0 Hz, H-15β) | ||
| 16 | 29.9 | 1.61 (1H, dddd, J = 13.8, 12.0, 8.4, 3.6 Hz, H-16α) |
| 2.09 (1H, dddd, J = 13.8, 9.6, 9.0, 6.0 Hz, H-16β) | ||
| 17 | 89.7 | 3.79 (1H, dd, J = 9.0, 8.4 Hz, H-17) |
| 18 | 12.0 | 0.90 (3H, s, H-18) |
| 19 | 17.7 | 1.24 (3H, s, H-19) |
| 1ʹ | 104.8 | 4.44 (1H, d, J = 7.8 Hz, H-1ʹ) |
| 2ʹ | 75.4 | 3.21 (1H, dd, J =9.0, 7.8 Hz H-2ʹ) |
| 3ʹ | 77.1 | 3.29 (1H, t, J = 9.0 Hz, H-3ʹ) |
| 4ʹ | 71.6 | 3.36 (1H, dd, J = 9.0, 8.4 Hz, H-4ʹ) |
| 5ʹ | 78.0 | 3.37 (1H, dd, J = 8.4, 5.4 Hz, H-5ʹ) |
| 6ʹ | 69.7 | 4.12 (1H, dd, J = 12.0, 2.4 Hz, H-6ʹa) |
| 3.79 (1H, dd, J = 12.0, 5.4 Hz, H-6ʹb) | ||
| 1″ | 104.8 | 4.35 (1H, d, J = 7.8 Hz, H-1″) |
| 2″ | 75.1 | 3.21 (1H, dd, J =9.0, 7.8 Hz, H-2″) |
| 3″ | 77.1 | 3.29 (1H, t, J = 9.0 Hz, H-3″) |
| 4″ | 71.5 | 3.41 (1H, dd, J = 9.0, 8.4 Hz, H- 4″) |
| 5″ | 78.0 | 3.37 (1H, dd, J = 8.4, 5.4 Hz, H-5″) |
| 6″ | 62.8 | 3.67 (1H, dd, J = 12.0, 2.4 Hz, H-6″a) |
| 3.88 (1H, dd, J = 12.0, 5.4 Hz, H-6″b) | ||
| No | Steroid | Reactivity |
|---|---|---|
| 1 | testosterone | + |
| 2 | deoxycorticosterone | + |
| 3 | hydrocortisone | + |
| 4 | estradiol | + |
| 5 | 4-androstenediol | + |
| 6 | 5-androstenediol | - |
| 7 | epitestosterone | - |
| 8 | cerberigenin | - |
| 9 | dehydroepiandrosterone | - |
| 10 | pregnenolone | - |
| 11 | 17α-hydroxypregnenolone | - |
| 12 | ethinyl estradiol | - |
| 13 | estrone | - |
| 14 | ethisterone | - |
| 15 | 17α-hydroxyprogesterone | - |
| 16 | 24-epicastasterone | - |
| 17 | diosgenin | - |
| 18 | ergosta-5,24(28)-diene-3,7,16-triol | - |
| 19 | cyasterone | - |
| 20 | cholesterol | - |
| 21 | β-sitosterol | - |
| 22 | ergosterol | - |
| 23 | campesterol | - |
| 24 | cholic acid | - |
| 25 | 11β-hydroxyprogesterone | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.-L.; Kong, J.-Q. OcUGT1-Catalyzing Glycodiversification of Steroids through Glucosylation and Transglucosylation Actions. Molecules 2020, 25, 475. https://doi.org/10.3390/molecules25030475
Xu Y-L, Kong J-Q. OcUGT1-Catalyzing Glycodiversification of Steroids through Glucosylation and Transglucosylation Actions. Molecules. 2020; 25(3):475. https://doi.org/10.3390/molecules25030475
Chicago/Turabian StyleXu, Yan-Li, and Jian-Qiang Kong. 2020. "OcUGT1-Catalyzing Glycodiversification of Steroids through Glucosylation and Transglucosylation Actions" Molecules 25, no. 3: 475. https://doi.org/10.3390/molecules25030475