Geographical Discrimination in Curcuminoids Content of Turmeric Assessed by Rapid UPLC-DAD Validated Analytical Method
Abstract
:1. Introduction
2. Results
2.1. Isolation and Purification of Curcuminoids
2.2. Optimization of Chromatographic Condition
2.3. Column Performance
2.4. Method Validation
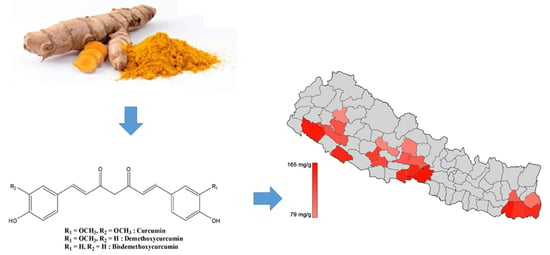
2.5. Quantification of Turmeric Samples
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Extraction Isolation and Identification of Curcuminoids
4.3. Chromatographic Condition
4.4. Column Performance
4.5. Preparation of Standard and Sample Solutions
4.6. Method Validation
4.7. Quantification of Turmeric Extractive Solution
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Xia, Q.; Zhao, K.J.; Huang, Z.G.; Zhang, P.; Dong, T.T.; Li, S.P.; Tsim, K.W. Molecular genetic and chemical assessment of Rhizoma Curcumae in China. J. Agric. Food Chem. 2005, 53, 6019–6026. [Google Scholar] [CrossRef] [PubMed]
- Dosoky, N.S.; Setzer, W.N. Chemical Composition and Biological Activities of Essential Oils of Curcuma Species. Nutrients 2018, 10, 1196. [Google Scholar] [CrossRef]
- Aggarwal, B.B.; Yuan, W.; Li, S.; Gupta, S.C. Curcumin-free turmeric exhibits anti-inflammatory and anticancer activities: Identification of novel components of turmeric. Mol. Nutr. Food Res. 2013, 57, 1529–1542. [Google Scholar] [CrossRef] [PubMed]
- Fadus, M.C.; Lau, C.; Bikhchandani, J.; Lynch, H.T. Curcumin: An age-old anti-inflammatory and anti-neoplastic agent. J. Tradit. Complement. Med. 2017, 7, 339–346. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Lim, G.P.; Begum, A.N.; Ubeda, O.J.; Simmons, M.R.; Ambegaokar, S.S.; Chen, P.P.; Kayed, R.; Glabe, C.G.; Frautschy, S.A.; et al. Curcumin inhibits formation of amyloid beta oligomers and fibrils, binds plaques, and reduces amyloid in vivo. J. Biol. Chem. 2005, 280, 5892–5901. [Google Scholar] [CrossRef] [PubMed]
- Serafini, M.M.; Catanzaro, M.; Rosini, M.; Racchi, M.; Lanni, C. Curcumin in Alzheimer’s disease: Can we think to new strategies and perspectives for this molecule? Pharmacol. Res. 2017, 124, 146–155. [Google Scholar] [CrossRef] [PubMed]
- Morales, I.; Cerda-Troncoso, C.; Andrade, V.; Maccioni, R.B. The Natural Product Curcumin as a Potential Coadjuvant in Alzheimer’s Treatment. J. Alzheimer’s Dis. JAD 2017, 60, 451–460. [Google Scholar] [CrossRef]
- Cheng, J.; Weijun, K.; Yun, L.; Jiabo, W.; Haitao, W.; Qingmiao, L.; Xiaohe, X. Development and validation of UPLC method for quality control of Curcuma longa Linn.: Fast simultaneous quantitation of three curcuminoids. J. Pharm. Biomed. Anal. 2010, 53, 43–49. [Google Scholar] [CrossRef]
- Zhang, J.; Wider, B.; Shang, H.; Li, X.; Ernst, E. Quality of herbal medicines: Challenges and solutions. Complement. Ther. Med. 2012, 20, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Saini, G.; Nair, A.; Sharma, R. UPLC: A preeminent technique in pharmaceutical analysis. Acta Pol. Pharm. 2012, 69, 371–380. [Google Scholar]
- Sandur, S.K.; Pandey, M.K.; Sung, B.; Ahn, K.S.; Murakami, A.; Sethi, G.; Limtrakul, P.; Badmaev, V.; Aggarwal, B.B. Curcumin, demethoxycurcumin, bisdemethoxycurcumin, tetrahydrocurcumin and turmerones differentially regulate anti-inflammatory and anti-proliferative responses through a ROS-independent mechanism. Carcinogenesis 2007, 28, 1765–1773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jayaprakasha, G.K.; Nagana Gowda, G.A.; Marquez, S.; Patil, B.S. Rapid separation and quantitation of curcuminoids combining pseudo two-dimensional liquid flash chromatography and NMR spectroscopy. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2013, 937, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Yadav, D.; Reshi, M.S.; Uthra, C.; Shrivastava, S.; Srivastava, N.; Narayana, S.K.K.; Shukla, S. Botanical and Chemical Fingerprinting of Medicinal Roots of Justicia gendarussa Burm f. Pharmacogn. Res. 2017, 9, 208–214. [Google Scholar]
- Ang, L.F.; Yam, M.F.; Fung, Y.T.; Kiang, P.K.; Darwin, Y. HPLC method for simultaneous quantitative detection of quercetin and curcuminoids in traditional chinese medicines. J. Pharmacopuncture 2014, 17, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Lee, H.J.; Shin, Y. Optimization and validation of high-performance liquid chromatography method for individual curcuminoids in turmeric by heat-refluxed extraction. J. Agric. Food Chem. 2013, 61, 10911–10918. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Ahumada Mdel, C.; Timmermann, B.N.; Gang, D.R. Biosynthesis of curcuminoids and gingerols in turmeric (Curcuma longa) and ginger (Zingiber officinale): Identification of curcuminoid synthase and hydroxycinnamoyl-CoA thioesterases. Phytochemistry 2006, 67, 2017–2029. [Google Scholar] [CrossRef]
- Feng, G.H.; Leonard, T.J. Characterization of the polyketide synthase gene (pksL1) required for aflatoxin biosynthesis in Aspergillus parasiticus. J. Bacteriol. 1995, 177, 6246–6254. [Google Scholar] [CrossRef]
- Feng, G.H.; Leonard, T.J. Culture conditions control expression of the genes for aflatoxin and sterigmatocystin biosynthesis in Aspergillus parasiticus and A. nidulans. Appl. Environ. Microbiol. 1998, 64, 2275–2277. [Google Scholar]
- Kiuchi, F.; Goto, Y.; Sugimoto, N.; Akao, N.; Kondo, K.; Tsuda, Y. Nematocidal activity of turmeric: Synergistic action of curcuminoids. Chem. Pharm. Bull. 1993, 41, 1640–1643. [Google Scholar] [CrossRef]
- Shabir, G.A. Validation of high-performance liquid chromatography methods for pharmaceutical analysis. Understanding the differences and similarities between validation requirements of the US Food and Drug Administration, the US Pharmacopeia and the International Conference on Harmonization. J. Chromatogr. A 2003, 987, 57–66. [Google Scholar]
- Poudel, A.; Kim, S.G.; Lamichhane, R.; Kim, Y.K.; Jo, H.K.; Jung, H.J. Quantitative assessment of traditional Oriental herbal formulation Samhwangsasim-tang using UPLC technique. J. Chromatogr. Sci. 2014, 52, 176–185. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds as well as turmeric are available from the authors. |






| Column | Standards | RT | N | K’ | α | TF | Rs |
|---|---|---|---|---|---|---|---|
| Polar Advantage II | Cur | 0.701 | 6476.13 | 1.93 | 1.25 | ||
| DMCur | 0.800 | 8260.23 | 2.27 | 1.18 | 1.15 | 2.81 | |
| BDMCur | 0.902 | 8883.97 | 2.90 | 1.27 | 1.13 | 3.10 | |
| Aquity BEH | Cur | 2.012 | 12227.65 | 2.32 | 1.12 | 1.29 | 2.11 |
| DMCur | 1.857 | 10798.41 | 2.06 | 1.13 | 1.30 | 2.04 | |
| BDMCur | 1.709 | 9397.53 | 1.82 | 1.45 | |||
| Triart ExRS | Cur | 1.775 | 9911.04 | 3.15 | 1.16 | 1.25 | 2.63 |
| DMCur | 1.586 | 8282.66 | 2.71 | 1.17 | 1.27 | 2.45 | |
| BDMCur | 1.414 | 6965.46 | 2.30 | 1.47 | |||
| Kinetex | Cur | 1.876 | 9579.02 | 2.49 | 1.12 | 1.22 | 1.91 |
| DMCur | 1.728 | 8547.42 | 2.21 | 1.13 | 1.20 | 1.82 | |
| BDMCur | 1.590 | 7285.05 | 1.96 | 1.20 | |||
| Modifier | |||||||
| Without Acid | Cur | 0.701 | 6476.13 | 1.93 | 1.25 | ||
| DMCur | 0.800 | 8260.23 | 2.27 | 1.18 | 1.15 | 2.81 | |
| BDMCur | 0.902 | 8883.97 | 2.90 | 1.27 | 1.13 | 3.10 | |
| Formic acid | Cur | 0.682 | 5349.52 | 1.88 | 1.28 | ||
| DMCur | 0.786 | 7304.67 | 2.29 | 1.22 | 1.17 | 2.62 | |
| BDMCur | 0.899 | 7809.62 | 2.79 | 1.21 | 1.13 | 2.95 | |
| Acetic acid | Cur | 0.668 | 5564.26 | 1.91 | 1.21 | ||
| DMCur | 0.767 | 7481.07 | 2.34 | 1.23 | 1.11 | 2.71 | |
| BDMCur | 0.883 | 8325.92 | 2.84 | 1.22 | 1.05 | 3.06 | |
| Trifluoroacetic acid | Cur | 0.660 | 5583.52 | 1.84 | 1.17 | ||
| DMCur | 0.751 | 7472.48 | 2.24 | 1.21 | 1.09 | 2.55 | |
| BDMCur | 0.859 | 8006.48 | 2.70 | 1.21 | 1.05 | 2.90 |
| Compound | Regression Equation † | R2 | LOD | LOQ |
|---|---|---|---|---|
| Curcumin | y = 35831x + 19962 | 0.999 | 0.155 | 0.470 |
| Demethoxycurcumin | y = 36541x + 26194 | 0.999 | 0.170 | 0.517 |
| Bisdemethoxycurcumin | y = 35536x + 13556 | 0.999 | 0.190 | 0.577 |
| Standards | Concentration | Intra-Day | Inter-Day | ||
|---|---|---|---|---|---|
| (μg/mL) | Mean ± SD (μg/mL) | RSD (%) | Mean ± SD (μg/mL) | RSD (%) | |
| Cur | 62.500 | 63.40 ± 0.10 | 0.16 | 63.76 ± 0.31 | 0.48 |
| 31.250 | 33.14 ± 0.17 | 0.51 | 33.34 ± 0.23 | 0.68 | |
| 15.625 | 16.29 ± 0.06 | 0.34 | 16.46 ± 0.16 | 0.95 | |
| DMCur | 31.250 | 32.41 ± 0.24 | 0.73 | 32.50 ± 0.35 | 1.08 |
| 15.625 | 17.29 ± 0.01 | 0.08 | 17.43 ± 0.17 | 0.95 | |
| 7.813 | 8.53 ± 0.02 | 0.26 | 8.67 ± 0.18 | 2.13 | |
| BDMCur | 7.813 | 7.95 ± 0.01 | 0.07 | 8.07 ± 0.11 | 1.31 |
| 3.906 | 4.03 ± 0.01 | 0.17 | 4.05 ± 0.03 | 0.68 | |
| 1.953 | 1.85 ± 0.01 | 0.32 | 1.88 ± 0.05 | 2.47 | |
| Standards | Amount ± SD (μg/mL) | RSD |
|---|---|---|
| Curcumin | 30.02 ± 0.12 | 0.39 |
| Demethoxycurcumin | 8.45 ± 0.02 | 0.26 |
| Bisdemethoxycurcumin | 3.14 ± 0.08 | 2.47 |
| Standards | Original (mg/mL) | Spiked (mg/mL) | Found ± SD (mg/mL) | Recovery (%) | RSD (%) |
|---|---|---|---|---|---|
| Cur | 25.49 | 7.81 | 7.82 ± 0.08 | 100.07 | 1.02 |
| 3.9 | 3.97 ± 0.11 | 101.80 | 2.79 | ||
| 1.95 | 2.02 ± 0.02 | 103.40 | 0.81 | ||
| DMCur | 13.17 | 7.81 | 7.94 ± 0.01 | 101.64 | 0.07 |
| 3.9 | 3.89 ± 0.00 | 99.87 | 0.12 | ||
| 1.95 | 1.91 ± 0.01 | 98.97 | 0.28 | ||
| BDMCur | 19.08 | 7.81 | 7.70 ± 0.09 | 98.54 | 1.12 |
| 3.9 | 4.05 ± 0.08 | 103.91 | 2.06 | ||
| 1.95 | 1.97 ± 0.01 | 101.12 | 0.55 |
| Longitude(N)/Latitude(E) | Altitude (m) | Cur (mg/g) | DMCur (mg/g) | BDMCur (mg/g) | |
|---|---|---|---|---|---|
| Korea Samples | |||||
| Jeju | 33.48/126.49 | 40 | 48.83 ± 0.12 | 14.01 ± 0.11 | 5.53 ± 0.05 |
| Jindo | 34.46/126.24 | 90 | 37.48 ± 0.34 | 10.38 ± 0.28 | 4.25 ± 0.06 |
| Koksong | 35.28/127.29 | 280 | 35.85 ± 0.05 | 9.43 ± 0.16 | 3.73 ± 0.08 |
| Nepal Samples | |||||
| Chitwan | 27.52/84.35 | 80 | 165.34 ± 0.08 | 65.34 ± 0.26 | 51.54 ± 0.78 |
| Sunsari | 26.62/87.18 | 100 | 165.12 ± 0.24 | 86.96 ± 0.20 | 66.83 ± 0.03 |
| Dhangadi | 28.68/80.62 | 109 | 159.80 ± 0.02 | 67.07 ± 0.27 | 95.33 ± 0.65 |
| Nawalparasi | 27.64/83.88 | 189 | 150.75 ± 0.69 | 66.48 ± 0.11 | 80.58 ± 0.16 |
| Banke | 28.14/81.77 | 215 | 148.08 ± 0.50 | 70.31 ± 0.14 | 65.23 ± 0.16 |
| Jhapa | 26.63/87.89 | 280 | 142.01 ± 0.37 | 73.26 ± 0.14 | 80.41 ± 0.45 |
| Morang | 26.67/87.46 | 400 | 135.26 ± 0.64 | 65.95 ± 0.25 | 86.85 ± 0.19 |
| Pyuthan | 28.10/82.85 | 450 | 132.21 ± 0.37 | 54.83 ± 0.26 | 44.48 ± 0.17 |
| Arghakanchi | 27.98/83.03 | 525 | 124.93 ± 2.46 | 59.88 ± 1.60 | 64.47 ± 2.20 |
| Surkhet | 28.51/81.77 | 750 | 119.66 ± 0.01 | 68.52 ± 0.08 | 51.38 ± 0.06 |
| Dailekh | 28.92/81.64 | 800 | 119.60 ± 0.07 | 64.10 ± 0.26 | 14.44 ± 0.29 |
| Syangja | 28.01/83.80 | 900 | 116.77 ± 0.10 | 79.07 ± 0.01 | 70.35 ± 0.30 |
| Tanahu | 27.94/84.22 | 988 | 115.39 ± 0.08 | 62.53 ± 0.06 | 85.24 ± 0.13 |
| Dhankuta | 26.98/87.32 | 998 | 114.89 ± 0.25 | 50.81 ± 0.09 | 50.52 ± 0.22 |
| Illam | 26.87/87.93 | 1060 | 103.94 ± 0.06 | 64.63 ± 0.62 | 116.09 ± 3.09 |
| Kaski | 28.26/84.01 | 1530 | 91.57 ± 0.27 | 63.75 ± 0.53 | 109.25 ± 0.75 |
| Baglung | 28.27/83.58 | 1750 | 80.28 ± 0.23 | 36.92 ± 0.09 | 43.95 ± 0.19 |
| Kalikot | 29.20/81.73 | 2150 | 79.14 ± 0.10 | 30.87 ± 0.11 | 32.07 ± 0.07 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poudel, A.; Pandey, J.; Lee, H.-K. Geographical Discrimination in Curcuminoids Content of Turmeric Assessed by Rapid UPLC-DAD Validated Analytical Method. Molecules 2019, 24, 1805. https://doi.org/10.3390/molecules24091805
Poudel A, Pandey J, Lee H-K. Geographical Discrimination in Curcuminoids Content of Turmeric Assessed by Rapid UPLC-DAD Validated Analytical Method. Molecules. 2019; 24(9):1805. https://doi.org/10.3390/molecules24091805
Chicago/Turabian StylePoudel, Amrit, Jitendra Pandey, and Hyeong-Kyu Lee. 2019. "Geographical Discrimination in Curcuminoids Content of Turmeric Assessed by Rapid UPLC-DAD Validated Analytical Method" Molecules 24, no. 9: 1805. https://doi.org/10.3390/molecules24091805