Novel Imidazole Aldoximes with Broad-Spectrum Antimicrobial Potency against Multidrug Resistant Gram-Negative Bacteria
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Biological Evaluation
2.2.1. Antimicrobial Activity
2.2.2. Activity toward Gram-Negative β-Lactamase Producing Strains
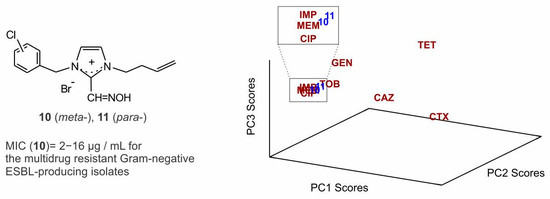
2.3. Principal Component Analysis
2.4. Combination Study of Compound 10 with Ceftazidime
3. Materials and Methods
3.1. General Information
3.2. Disc Diffusion Assay
3.3. Minimum Inhibitory Concentration Assay
3.4. Principal Component Analysis
4. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- The Global Risks Report 2017, 12th ed.; World Economic Forum: Geneva, Switzerland, 2017; pp. 1–78. ISBN 978-1-944835-07-1.
- Boucher, H.W.; Talbot, G.H.; Benjamin, D.K., Jr.; Bradley, J.; Guidos, R.J.; Jones, R.N.; Murray, B.E.; Bonomo, R.A.; Gilbert, D. 10 × ′20 Progress—development of new drugs active against gram-negative bacilli: An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2013, 56, 1685–1694. [Google Scholar] [CrossRef] [PubMed]
- Bush, K.; Jacoby, G.A. Updated functional classification of beta-lactamases. Antimicrob. Agents Chemother. 2010, 54, 969–976. [Google Scholar] [CrossRef] [PubMed]
- Canton, R.; Coque, T.M. The CTX-M beta-lactamase pandemic. Curr. Opin. Microbiol. 2006, 9, 466–475. [Google Scholar] [CrossRef] [PubMed]
- Drawz, S.M.; Bonomo, R.A. Three Decades of β-Lactamase Inhibitors. Clin. Microbiol. Rev. 2010, 23, 160–201. [Google Scholar] [CrossRef] [PubMed]
- Livermore, D.M.; Mushtaq, S.; Warner, M.; Zhang, J.; Maharjan, S.; Doumith, M.; Woodford, N. Activities of NXL104 Combinations with Ceftazidime and Aztreonam against Carbapenemase-Producing Enterobacteriaceae. Antimicrob. Agents Chemother. 2011, 55, 390–394. [Google Scholar] [CrossRef] [PubMed]
- Lagacé-Wiens, P.R.S.; Tailor, F.; Simner, P.; DeCorby, M.; Karlowsky, J.A.; Walkty, A.; Hoban, D.J.; Zhanel, G.G. Activity of NXL104 in Combination with β-Lactams against Genetically Characterized Escherichia coli and Klebsiella pneumoniae Isolates Producing Class A Extended-Spectrum β-Lactamases and Class C β-Lactamases. Antimicrob. Agents Chemother. 2011, 55, 2434–2437. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Peng, X.M.; Damu, G.L.; Geng, R.X.; Zhou, C.H. Comprehensive review in current developments of imidazole-based medicinal chemistry. Med. Res. Rev. 2014, 34, 340–437. [Google Scholar] [CrossRef] [PubMed]
- Boiani, M.; Gonzalez, M. Imidazole and benzimidazole derivatives as chemotherapeutic agents. Mini Rev. Med. Chem. 2005, 5, 409–424. [Google Scholar] [CrossRef] [PubMed]
- De Luca, L. Naturally occurring and synthetic imidazoles: Their chemistry and their biological activities. Curr. Med. Chem. 2006, 13, 1–23. [Google Scholar] [PubMed]
- Gaba, M.; Mohan, C. Development of drugs based on imidazole and benzimidazole bioactive heterocycles: Recent advances and future directions. Med. Chem. Res. 2016, 25, 173–210. [Google Scholar] [CrossRef]
- Dong, J.; Chen, S.; Li, R.; Cui, W.; Jiang, H.; Ling, Y.; Yang, Z.; Hu, W. Imidazole-based pinanamine derivatives: Discovery of dual inhibitors of the wild-type and drug-resistant mutant of the influenza A virus. Eur. J. Med. Chem. 2016, 108, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Seyedmousavi, S.; Rafati, H.; Ilkit, M.; Tolooe, A.; Hedayati, M.T.; Verweij, P. Systemic Antifungal Agents: Current Status and Projected Future Developments. Methods Mol. Biol. 2017, 1508, 107–139. [Google Scholar] [PubMed]
- Olekhnovich, I.N.; Vitko, S.; Valliere, M.; Hoffman, P.S. Response to Metronidazole and Oxidative Stress Is Mediated through Homeostatic Regulator HsrA (HP1043) in Helicobacter pylori. J. Bacteriol. 2014, 196, 729–739. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Sun, H.; Zhang, Y.; Mukkamala, D.; Oldfield, E. A Solid State 13C NMR, Crystallographic, and Quantum Chemical Investigation of Chemical Shifts and Hydrogen Bonding in Histidine Dipeptides. J. Am. Chem. Soc. 2005, 127, 12544–12554. [Google Scholar] [CrossRef] [PubMed]
- Bunnage, M.E.; Blagg, J.; Steele, J.; Owen, D.R.; Allerton, C.; McElroy, A.B.; Miller, D.; Ringer, T.; Butcher, K.; Beaumont, K.; et al. Discovery of potent & selective inhibitors of activated thrombin-activatable fibrinolysis inhibitor for the treatment of thrombosis. J. Med. Chem. 2007, 50, 6095–6103. [Google Scholar] [PubMed]
- Salerno, L.; Pittala, V.; Romeo, G.; Modica, M.N.; Marrazzo, A.; Siracusa, M.A.; Sorrenti, V.; Di Giacomo, C.; Vanella, L.; Parayath, N.N.; et al. Novel imidazole derivatives as heme oxygenase-1 (HO-1) and heme oxygenase-2 (HO-2) inhibitors and their cytotoxic activity in human-derived cancer cell lines. Eur. J. Med. Chem. 2015, 96, 162–172. [Google Scholar] [CrossRef] [PubMed]
- Silva, V.G.; Silva, R.O.; Damasceno, S.R.; Carvalho, N.S.; Prudencio, R.S.; Aragao, K.S.; Guimarães, M.A.; Campos, S.A.; Véras, L.M.; Godejohann, M.; et al. Anti-inflammatory and antinociceptive activity of epiisopiloturine, an imidazole alkaloid isolated from Pilocarpus microphyllus. J. Nat. Prod. 2013, 76, 1071–1077. [Google Scholar] [CrossRef] [PubMed]
- Shelton, K.L.; DeBord, M.A.; Wagers, P.O.; Southerland, M.R.; Taraboletti, A.; Robishaw, N.K.; Jackson, D.P.; Tosanovic, R.; Kofron, W.G.; Tessier, C.A.; et al. Synthesis, anti-proliferative activity, and toxicity of C4(C5) substituted N,N′-bis(arylmethyl)imidazolium salts. Tetrahedron 2016, 72, 5729–5743. [Google Scholar] [CrossRef]
- Zhao, D.M.; Zhao, S.Z.; Zhao, L.Y.; Zhang, X.Q.; Wei, P.; Liu, C.C.; Hao, C.; Sun, B.; Su, X.; Cheng, M. Discovery of biphenyl imidazole derivatives as potent antifungal agents: Design, synthesis, and structure-activity relationship studies. Bioorg. Med. Chem. 2017, 25, 750–758. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulou, M.V.; Bloomer, W.D.; Rosenzweig, H.S.; Arena, A.; Arrieta, F.; Rebolledo, J.C.; Smith, D.K. Nitrotriazole- and imidazole-based amides and sulfonamides as antitubercular agents. Antimicrob. Agents Chemother. 2014, 58, 6828–6836. [Google Scholar] [CrossRef] [PubMed]
- Kantsadi, A.L.; Bokor, E.; Kun, S.; Stravodimos, G.A.; Chatzileontiadou, D.S.M.; Leonidas, D.D.; Juhász-Tóth, É.; Szakács, A.; Batta, G.; Docsa, T.; et al. Synthetic, enzyme kinetic, and protein crystallographic studies of C-beta-d-glucopyranosyl pyrroles and imidazoles reveal and explain low nanomolar inhibition of human liver glycogen phosphorylase. Eur. J. Med. Chem. 2016, 123, 737–745. [Google Scholar] [CrossRef] [PubMed]
- Gong, K.K.; Tang, X.L.; Liu, Y.S.; Li, P.L.; Li, G.Q. Imidazole Alkaloids from the South China Sea Sponge Pericharax heteroraphis and Their Cytotoxic and Antiviral Activities. Molecules 2016, 21, 150. [Google Scholar] [CrossRef] [PubMed]
- Odžak, R.; Skočibušić, M.; Maravić, A. Synthesis and antimicrobial profile of N-substituted imidazolium oximes and their monoquaternary salts against multidrug resistant bacteria. Bioorg. Med. Chem. 2013, 21, 7499–7506. [Google Scholar]
- Sugiyama, Y.; Ohtani, I.I.; Isobe, M.; Takai, A.; Ubukata, M.; Isono, K. Molecular shape analysis and activity of tautomycin, a protein phosphatase inhibitor. Bioorg. Med. Chem. Lett. 1996, 6, 3–8. [Google Scholar] [CrossRef]
- Quinn, D.M. Acetylcholinesterase: Enzyme structure, reaction dynamics, and virtual transition states. Chem. Rev. 1987, 87, 955–979. [Google Scholar] [CrossRef]
- Simeon-Rudolf, V.; Reiner, E.; Škrinjarić-Špoljar, M.; Radić, B.; Lucić, A.; Primožič, I.; Tomić, S. Quinuclidinium-imidazolium compounds: Synthesis, mode of interaction with acetylcholinesterase and effect upon Soman intoxicated mice. Arch. Toxicol. 1998, 72, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Iversen, P.E.; Lund, H. Preparation of 2-imidazole- and 2-thiazolecarbaldehydes. Acta Chem. Scand. 1966, 20, 2649–2657. [Google Scholar] [CrossRef]
- Katalinić, M.; Maček Hrvat, N.; Baumann, K.; Morasi Piperčić, S.; Makarić, S.; Tomić, S.; Jović, O.; Hrenar, T.; Miličević, A.; Jelić, D.; et al. A comprehensive evaluation of novel oximes in creation of butyrylcholinesterase-based nerve agent bioscavengers. Toxicol. Appl. Pharmacol. 2016, 310, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Mena, A.; Smith, E.E.; Burns, J.L.; Speert, D.P.; Moskowitz, S.M.; Perez, J.L.; Oliver, A. Genetic adaptation of Pseudomonas aeruginosa to the airways of cystic fibrosis patients is catalyzed by hypermutation. J. Bacteriol. 2008, 190, 7910–7917. [Google Scholar] [CrossRef] [PubMed]
- Beltrami, E. Sulle funzioni bilineari. In Giornale di Matematiche ad Uso degli Studenti Delle Universita; Benedetto Pellerano Editore: Napoli, Italy, 1873; Volume 11, pp. 98–106. [Google Scholar]
- Pearson, K. On lines and planes of closest fit to systems of points in space. Philos. Mag. 1901, 2, 559–572. [Google Scholar] [CrossRef]
- Hotelling, H. Analysis of a Complex of Statistical Variables into Principal Components. J. Educ. Psychol. 1933, 24, 417–441. [Google Scholar] [CrossRef]
- Jolliffe, I.T. Principal Component Analysis; Springer: Berlin, Germany, 1986. [Google Scholar]
- Smilde, A.; Bro, R.; Geladi, P. Multi-Way Analysis with Applications in the Chemical Sciences; John Wiley & Sons Ltd.: Chichester, UK, 2004. [Google Scholar]
- Hrenar, T. moonee, Program for Manipulation and Analysis of Multi- and Univariate Data, rev. 0.6827. 2018. [Google Scholar]
- Geladi, P.; Kowalski, B. Partial least-squares regression, a tutorial. Anal. Chim. Acta 1986, 185, 1–17. [Google Scholar] [CrossRef]
Sample Availability: Not available. |




| Microorganisms | Diameters of the Inhibition Zone (mm) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Compound No a | Antibiotic b | |||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | GEN | |
| Gram-positive bacteria | ||||||||||||
| Bacillus cereus | 16.4 ± 0.3 | 15.4 ± 0.5 | 11.0 ± 0.5 | 13.3 ± 0.8 | 13.6 ± 0.8 | 19.2 ± 0.5 | 16.2 ± 0.2 | 19.1 ± 0.6 | 12.1 ± 0.6 | 21.2 ± 0.6 | 13.6 ± 0.1 | 18.2 ± 0.7 |
| Enterococcus faecalis | 17.7 ± 1.2 | 19.2 ± 0.4 | 16.2 ± 0.6 | 10.7 ± 0.6 | 12.7 ± 0.5 | 14.7 ± 0.7 | 20.4 ± 0.6 | 12.3 ± 0.7 | 20.3 ± 0.4 | 16.3 ± 0.2 | 14.5 ± 0.7 | 14.6 ± 1.4 |
| Staphylococcus aureus | 16.1 ± 0.9 | 15.1 ± 0.2 | 14.4 ± 0.2 | 12.3 ± 0.5 | 16.3 ± 2.4 | 16.1 ± 0.5 | 15.6 ± 1.6 | 19.7 ± 0.3 | 20.8 ± 0.7 | 16.7 ± 0.9 | 12.1 ± 0.8 | 23.9 ± 0.9 |
| Clostridium perfringens | 15.5 ± 0.2 | 11.7 ± 0.6 | 17.6 ±1.4 | 11.2 ± 0.8 | 14.4 ± 1.1 | 13.9 ± 1.2 | 17.7 ± 0.9 | 19.4 ± 0.4 | 15.2 ± 0.3 | 14.1 ± 1.0 | 15.9 ± 0.2 | 21.7 ± 0.4 |
| Gram-negative bacteria | ||||||||||||
| Escherichia coli | 18.8 ± 0.3 | 18.9 ± 0.2 | 21.8 ± 0.9 | 17.3 ± 1.1 | 18.3 ± 0.4 | 20.7 ± 0.5 | 17.1 ± 0.9 | 20.7 ± 0.8 | 12.4 ± 0.7 | 20.4 ± 1.1 | 13.4 ± 0.7 | 11.5 ± 0.9 |
| Klebsiella pneumoniae | 15.2 ± 1.5 | 16.3 ± 0.1 | 17.3 ± 0.0 | 14.8 ± 0.3 | 16.5 ± 0.1 | 20.1 ± 0.8 | 17.5 ± 0.8 | 19.4 ± 0.2 | 14.1 ± 0.3 | 18.2 ± 0.8 | 16.8 ± 0.5 | 18.8 ± 0.6 |
| Pseudomonas aeruginosa | 16.8 ± 1.9 | 18.9 ± 0.4 | 16.8 ± 0.3 | 14.9 ± 0.1 | 14.9 ± 0.3 | 20.2 ± 0.4 | 18.6 ± 0.4 | 16.7 ± 04 | 13.3 ± 0.7 | 18.8 ± 1.6 | 12.2 ± 1.1 | 9.7 ± 1.4 |
| Cronobacter sakazakii | 13.7 ± 1.5 | 15.7 ± 1.9 | 18.3 ± 0.1 | 13.1 ± 0.3 | 12.3 ± 0.8 | 13.6 ± 0.2 | 13.7 ± 0.1 | 16.1 ± 0.6 | 12.4 ± 0.1 | 16.4 ± 0.7 | 12.1 ± 0.6 | 13.6 ± 0.9 |
| MIC | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Compound No | Antibiotic | |||||||||||||||||||||||
| Microorganisms | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | GEN | ||||||||||||
| µg/mL | μmol/L | µg/mL | μmol/L | µg/mL | μmol/L | µg/mL | μmol/L | µg/mL | μmol/L | µg/mL | μmol/L | µg/mL | μmol/L | µg/mL | μmol/L | µg/mL | μmol/L | µg/mL | μmol/L | µg/mL | μmol/L | µg/mL | μmol/L | |
| Gram-positive bacteria | ||||||||||||||||||||||||
| Bacillus cereus | 25.00 | 163.2 | 50.00 | 299.0 | 50.00 | 302.7 | 25.00 | 106.1 | 25.00 | 106.1 | 12.50 | 34.9 | 25.00 | 69.7 | 25.00 | 67.1 | 25.00 | 67.1 | 25.00 | 67.5 | 12.50 | 33.7 | 4.00 | 8.4 |
| Enterococcus faecalis | 12.50 | 81.6 | 50.00 | 299.0 | 50.00 | 302.7 | 25.00 | 106.1 | 25.00 | 106.1 | 25.00 | 69.7 | 25.00 | 69.7 | 25.00 | 67.1 | 50.00 | 134.2 | 25.00 | 67.5 | 25.00 | 67.5 | 4.00 | 8.4 |
| Staphylococcus aureus | 50.00 | 326.4 | 25.00 | 149.5 | 25.00 | 151.3 | 12.50 | 53.0 | 25.00 | 106.1 | 25.00 | 69.7 | 12.50 | 34.9 | 25.00 | 67.1 | 50.00 | 134.2 | 25.00 | 67.5 | 50.00 | 134.9 | 1.00 | 2.1 |
| Clostridium perfringens | 25.00 | 163.2 | 12.50 | 74.8 | 25.00 | 151.3 | 25.00 | 106.1 | 12.50 | 53.0 | 50.00 | 139.4 | 12.50 | 34.9 | 25.00 | 67.1 | 50.00 | 134.2 | 25.00 | 67.5 | 12.50 | 33.7 | 0.50 | 1.1 |
| Gram-negative bacteria | ||||||||||||||||||||||||
| Escherichia coli | 12.50 | 81.6 | 25.00 | 149.5 | 50.00 | 302.7 | 12.50 | 53.0 | 50.00 | 212.2 | 25.00 | 69.7 | 25.00 | 69.7 | 50.00 | 134.2 | 50.00 | 134.2 | 6.25 | 16.9 | 25.00 | 67.5 | 32.00 | 67.0 |
| Klebsiella pneumoniae | 25.00 | 163.2 | 25.00 | 149.5 | 50.00 | 302.7 | 25.00 | 106.1 | 25.00 | 106.1 | 25.00 | 69.7 | 25.00 | 69.7 | 25.00 | 67.1 | 50.00 | 134.2 | 6.25 | 16.9 | 25.00 | 67.5 | 8.00 | 16.8 |
| Pseudomonas aeruginosa | 25.00 | 163.2 | 50.00 | 299.0 | 25.00 | 151.3 | 25.00 | 106.1 | 50.00 | 212.2 | 25.00 | 69.7 | 25.00 | 69.7 | 50.00 | 134.2 | 50.00 | 134.2 | 12.50 | 33.7 | 12.50 | 33.7 | 64.00 | 134.0 |
| Cronobacter sakazakii | 25.00 | 163.2 | 25.00 | 149.5 | 25.00 | 151.3 | 25.00 | 106.1 | 50.00 | 212.2 | 50.00 | 139.4 | 25.00 | 69.7 | 50.00 | 134.2 | 50.00 | 134.2 | 25.00 | 67.5 | 25.00 | 67.5 | 8.00 | 16.8 |
| Species, ESBL | MIC | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Compounds | Standard Antibiotics a | |||||||||||||||||||
| 10 | 11 | CAZ | CTX | CIP | GEN | TET | TOB | IMP | MEM | |||||||||||
| µg/mL | μmol/L | µg/mL | μmol/L | µg/mL | μmol/L | µg/mL | μmol/L | µg/mL | μmol/L | µg/mL | μmol/L | µg/mL | μmol/L | µg/mL | μmol/L | µg/mL | μmol/L | µg/mL | μmol/L | |
| Klebsiella pneumoniae, TEM-1, SHV-11, CTX-15 | 4 | 10.8 | 12 | 32.4 | 128 | 234.2 | 64 | 140.5 | 2.00 | 6.0 | 32 | 67.0 | 256 | 576.0 | 2.0 | 4.3 | 0.12 | 0.4 | 0.12 | 0.3 |
| Klebsiella pneumoniae, TEM-1, CTX-15, AmpC | 12 | 32.4 | 32 | 86.3 | >128 | >234.2 | >256 | >562.1 | 0.25 | 0.8 | 4 | 8.4 | 16 | 36.0 | 32 | 68.4 | 0.25 | 0.8 | 0.50 | 1.3 |
| Escherichia coli, CTX-15 | 8 | 21.6 | 16 | 43.2 | 8 | 14.6 | >256 | >562.1 | 0.50 | 1.5 | 8 | 16.8 | 128 | 288.0 | 64 | 136.9 | 0.12 | 0.4 | 1.00 | 2.6 |
| Escherichia coli, SHV-12, CTX-15 | 4 | 10.8 | 12 | 32.4 | 64 | 117.1 | >256 | >562.1 | 0.50 | 1.5 | 16 | 33.5 | 32 | 72.0 | 16 | 34.2 | 0.06 | 0.2 | 0.12 | 0.3 |
| Escherichia coli, TEM-1, SHV-12 | 2 | 5.4 | 12 | 32.4 | 16 | 29.3 | 32 | 70.3 | 1.00 | 3.0 | 128 | 268.0 | 64 | 144.0 | >32 | >68.4 | 8 | 26.7 | 0.25 | 0.7 |
| Enterobacter cloacae, SHV-12 | 12 | 32.4 | 4 | 10.8 | 8 | 14.6 | 8 | 17.6 | 0.50 | 1.5 | 2 | 4.2 | 16 | 36.0 | >32 | >68.4 | 0.12 | 0.4 | 0.12 | 0.3 |
| Enterobacter cloacae, TEM-1, SHV-12, CTX-15 | 2 | 5.4 | 8 | 21.6 | 64 | 117.1 | 256 | 562.1 | 2.00 | 6.0 | 4 | 8.4 | 8 | 18.0 | 16 | 34.2 | 0.06 | 0.2 | 0.06 | 0.2 |
| Raultella terrigena, TEM-1, SHV-11, CTX-15 | 4 | 10.8 | 32 | 86.3 | 4 | 7.3 | 256 | 562.1 | 0.50 | 1.5 | 128 | 268.0 | 64 | 144.0 | 32 | 68.4 | 0.25 | 0.8 | 0.125 | 0.3 |
| Citrobacter freundii, TEM-1, SHV-12, CTX-15, AmpC | 8 | 21.6 | 12 | 32.4 | 128 | 234.2 | 256 | 562.1 | 1.00 | 3.0 | 32 | 67.0 | 128 | 288.0 | 32 | 68.4 | 2.00 | 6.7 | 1.00 | 2.6 |
| Pantoea agglomerans, CTX-15 | 16 | 43.2 | 4 | 10.8 | 16 | 29.3 | 256 | 562.1 | 0.25 | 0.8 | 64 | 134.0 | 16 | 36.0 | 32 | 68.4 | 0.25 | 0.8 | 0.12 | 0.3 |
| Species, ESBL | MIC CAZ Alone | MIC 10 Alone | MIC Combination CAZ, 10 | |||
|---|---|---|---|---|---|---|
| (μg/mL) | (μmol/L) | (μg/mL) | (μmol/L) | (μg/mL) | (μmol/L) | |
| Klebsiella pneumoniae, TEM-1, SHV-11, CTX-15 | 128 | 234 | 4 | 11 | 4.8, 3.2 | 8.8, 8.6 |
| Klebsiella pneumoniae, TEM-1, CTX-15, AmpC | >128 | 234 | 12 | 32 | 9.6, 6.4 | 17.6, 17.3 |
| Escherichia coli, CTX-15 | 8 | 15 | 8 | 22 | 0.6, 0.4 | 1.1, 1.1 |
| Escherichia coli, SHV-12, CTX-15 | 64 | 117 | 4 | 11 | 0.3, 0.2 | 0.5, 0.5 |
| Escherichia coli, TEM-1, SHV-12 | 16 | 29 | 2 | 5 | 0.6, 0.4 | 1.1, 1.1 |
| Enterobacter cloacae, SHV-12 | 8 | 15 | 12 | 32 | 0.3, 0.2 | 0.5, 0.5 |
| Enterobacter cloacae, TEM-1, SHV-12, CTX-15 | 64 | 117 | 2 | 5 | 1.2, 0.8 | 2.2, 2.2 |
| Raultella terrigena, TEM-1, SHV-11, CTX-15 | 4 | 7 | 4 | 11 | 2.4, 1.6 | 4.4, 4.3 |
| Citrobacter freundii, TEM-1, SHV-12, CTX-15, AmpC | 128 | 234 | 8 | 22 | 4.8, 3.2 | 8.8, 8.6 |
| Pantoea agglomerans, CTX-15 | 16 | 29 | 16 | 43 | 0.6, 0.4 | 1.1, 1.1 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skočibušić, M.; Odžak, R.; Ramić, A.; Smolić, T.; Hrenar, T.; Primožič, I. Novel Imidazole Aldoximes with Broad-Spectrum Antimicrobial Potency against Multidrug Resistant Gram-Negative Bacteria. Molecules 2018, 23, 1212. https://doi.org/10.3390/molecules23051212
Skočibušić M, Odžak R, Ramić A, Smolić T, Hrenar T, Primožič I. Novel Imidazole Aldoximes with Broad-Spectrum Antimicrobial Potency against Multidrug Resistant Gram-Negative Bacteria. Molecules. 2018; 23(5):1212. https://doi.org/10.3390/molecules23051212
Chicago/Turabian StyleSkočibušić, Mirjana, Renata Odžak, Alma Ramić, Tomislav Smolić, Tomica Hrenar, and Ines Primožič. 2018. "Novel Imidazole Aldoximes with Broad-Spectrum Antimicrobial Potency against Multidrug Resistant Gram-Negative Bacteria" Molecules 23, no. 5: 1212. https://doi.org/10.3390/molecules23051212