Role of Gln79 in Feedback Inhibition of the Yeast γ-Glutamyl Kinase by Proline
Abstract
:1. Introduction
2. Materials and Methods
2.1. Yeast Strains, Media, and Plasmids
2.2. Isolation of AZC-Resistant Mutants from Awamori Yeast
2.3. Spot Test
2.4. Quantification of Intracellular Proline and Glutamate Contents
2.5. Homology Modeling of Pro1 and Docking Simulation of Proline into Pro1
2.6. Expression and Purification of Recombinant Pro1
2.7. Assay of GK Activity of Pro1
2.8. Western Blotting
2.9. Statistical Analysis
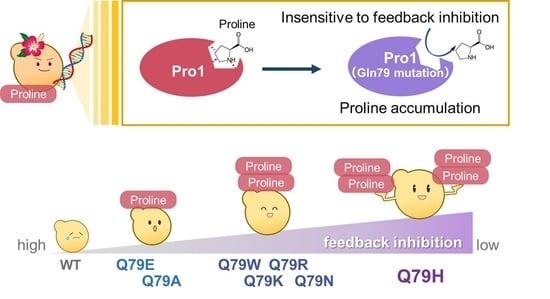
3. Results and Discussion
3.1. Isolation of Awamori Yeast with Intracellular Proline Accumulation
3.2. Characterization of the Q79H Variant of Pro1
3.3. Importance of Amino Acid Residue at Position 79 within Pro1
4. Summary
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murooka, Y.; Yamshita, M. Traditional healthful fermented products of Japan. J. Ind. Microbiol. Biotechnol. 2008, 35, 791–798. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Zhang, L.; Natarajan, S.K.; Becker, D.F. Proline mechanisms of stress survival. Antioxid. Redox Signal. 2013, 19, 998–1011. [Google Scholar] [CrossRef] [Green Version]
- Hayat, S.; Hayat, Q.; Alyemeni, M.N.; Wani, A.S.; Pichtel, J.; Ahmad, A. Role of proline under changing environments: A review. Plant Signal. Behav. 2012, 7, 1456–1466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Moukhtari, A.; Cabassa-Hourton, C.; Farissi, M.; Savouré, A. How does proline treatment promote salt stress tolerance during crop plant development? Front. Plant. Sci. 2020, 11, 1127. [Google Scholar] [CrossRef] [PubMed]
- Mat Nanyan, N.S.B.; Takagi, H. Proline homeostasis in Saccharomyces cerevisiae: How does the stress-responsive transcription factor Msn2 play a role? Front. Genet. 2020, 11, 438. [Google Scholar] [CrossRef] [PubMed]
- Rudolph, A.S.; Crowe, J.H. Membrane stabilization during freezing: The role of two natural cryoprotectants, trehalose and proline. Cryobiology 1985, 22, 367–377. [Google Scholar] [CrossRef]
- Solano, F. Metabolism and functions of amino acids in the skin. Adv. Exp. Med. Biol. 2020, 1265, 187–199. [Google Scholar]
- Signorelli, S.; Coitiño, E.L.; Borsani, O.; Monza, J. Molecular mechanisms for the reaction between •OH radicals and proline: Insights on the role as reactive oxygen species scavenger in plant stress. J. Phys. Chem. B 2014, 118, 37–47. [Google Scholar] [CrossRef]
- Tomenchok, D.M.; Brandriss, M.C. Gene-enzyme relationships in the proline biosynthetic pathway of Saccharomyces cerevisiae. J. Bacteriol. 1987, 169, 5364–5372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brandriss, M.C.; Falvey, D.A. Proline biosynthesis in Saccharomyces cerevisiae: Analysis of the PRO3 gene, which encodes delta 1-pyrroline-5-carboxylate reductase. J. Bacteriol. 1992, 174, 3782–3788. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.S.; Brandriss, M.C. Proline utilization in Saccharomyces cerevisiae: Analysis of the cloned PUT1 gene. Mol. Cell. Biol. 1986, 6, 2638–2645. [Google Scholar] [CrossRef] [Green Version]
- Brandriss, M.C. Proline utilization in Saccharomyces cerevisiae: Analysis of the cloned PUT2 gene. Mol. Cell. Biol. 1983, 3, 1846–1856. [Google Scholar] [CrossRef] [PubMed]
- Pandhare, J.; Donald, S.P.; Cooper, S.K.; Phang, J.M. Regulation and function of proline oxidase under nutrient stress. J. Cell. Biochem. 2009, 107, 759–768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sekine, T.; Kawaguchi, A.; Hamano, Y.; Takagi, H. Desensitization of feedback inhibition of the Saccharomyces cerevisiae gamma-glutamyl kinase enhances proline accumulation and freezing tolerance. Appl. Environ. Microbiol. 2007, 73, 4011–4019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takagi, H.; Matsui, F.; Kawaguchi, A.; Wu, H.; Shimoi, H.; Kubo, Y. Construction and analysis of self-cloning sake yeasts that accumulate proline. J. Biosci. Bioeng. 2007, 103, 377–380. [Google Scholar] [CrossRef]
- Fichman, Y.; Gerdes, S.Y.; Kovács, H.; Szabados, L.; Zilberstein, A.; Csonka, L.N. Evolution of proline biosynthesis: Enzymology, bioinformatics, genetics, and transcriptional regulation. Biol. Rev. 2015, 90, 1065–1099. [Google Scholar] [CrossRef]
- Pérez-Arellano, I.; Carmona-Álvarez, F.; Gallego, J.; Cervera, J. Molecular mechanisms modulating glutamate kinase activity. Identification of the proline feedback inhibitor binding site. J. Mol. Biol. 2010, 404, 890–901. [Google Scholar] [CrossRef]
- Marco-Marín, C.; Gil-Ortiz, F.; Pérez-Arellano, I.; Cervera, J.; Fita, I.; Rubio, V. A novel two-domain architecture within the amino acid kinase enzyme family revealed by the crystal structure of Escherichia coli glutamate 5-kinase. J. Mol. Biol. 2007, 367, 1431–1446. [Google Scholar] [CrossRef] [Green Version]
- Tatehashi, Y.; Takagi, H. Characterization of γ-glutamyl kinase mutants from Saccharomyces cerevisiae. J. Biosci. Bioeng. 2013, 116, 576–579. [Google Scholar] [CrossRef]
- Gietz, R.D.; Schiestl, R.H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007, 2, 31–34. [Google Scholar] [CrossRef]
- Nishimura, A.; Tanikawa, T.; Takagi, H. Inhibitory effect of arginine on proline utilization in Saccharomyces cerevisiae. Yeast 2020, 37, 531–540. [Google Scholar] [CrossRef]
- Trotter, E.W.; Kao, C.M.; Berenfeld, L.; Botstein, D.; Petsko, G.A.; Gray, J.V. Misfolded proteins are competent to mediate a subset of the responses to heat shock in Saccharomyces cerevisiae. J. Biol. Chem. 2002, 277, 44817–44825. [Google Scholar] [CrossRef] [Green Version]
- Terao, Y.; Nakamori, S.; Takagi, H. Gene dosage effect of L-proline biosynthetic enzymes on L-proline accumulation and freeze tolerance in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2003, 69, 6527–6532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takagi, H. Proline as a stress protectant in yeast: Physiological functions, metabolic regulations and biotechnological applications. Appl. Microbiol. Biotech. 2008, 81, 211–223. [Google Scholar] [CrossRef] [PubMed]
- Takagi, H. Metabolic regulatory mechanisms and physiological roles of functional amino acids and their applications in yeast. Biosci. Biotech. Biochem. 2019, 83, 1449–1462. [Google Scholar] [CrossRef] [PubMed]
- Takagi, H. Molecular mechanisms and highly-functional development for stress tolerance of the yeast Saccharomyces cerevisiae. Biosci. Biotech. Biochem. 2021, 85, 1017–1037. [Google Scholar] [CrossRef] [PubMed]
- Tsolmonbaatar, A.; Hashida, K.; Sugimoto, Y.; Watanabe, D.; Furukawa, S.; Takagi, H. Isolation of baker’s yeast mutants with proline accumulation that showed enhanced tolerance to baking-associated stresses. Int. J. Food Microbiol. 2016, 238, 233–240. [Google Scholar] [CrossRef] [PubMed]








| WT | Q79A | Q79E | Q79K | Q79N | Q79R | Q79W | |
|---|---|---|---|---|---|---|---|
| Specific activity (U/mg) | 17.38 ± 0.84 | 17.88 ± 3.88 | 2.18 ± 0.72 | 13.58 ± 1.97 | 19.13 ± 2.20 | 9.12 ± 1.04 | 18.36 ± 4.39 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nishimura, A.; Takasaki, Y.; Isogai, S.; Toyokawa, Y.; Tanahashi, R.; Takagi, H. Role of Gln79 in Feedback Inhibition of the Yeast γ-Glutamyl Kinase by Proline. Microorganisms 2021, 9, 1902. https://doi.org/10.3390/microorganisms9091902
Nishimura A, Takasaki Y, Isogai S, Toyokawa Y, Tanahashi R, Takagi H. Role of Gln79 in Feedback Inhibition of the Yeast γ-Glutamyl Kinase by Proline. Microorganisms. 2021; 9(9):1902. https://doi.org/10.3390/microorganisms9091902
Chicago/Turabian StyleNishimura, Akira, Yurie Takasaki, Shota Isogai, Yoichi Toyokawa, Ryoya Tanahashi, and Hiroshi Takagi. 2021. "Role of Gln79 in Feedback Inhibition of the Yeast γ-Glutamyl Kinase by Proline" Microorganisms 9, no. 9: 1902. https://doi.org/10.3390/microorganisms9091902