Next-Generation Probiotics and Their Metabolites in COVID-19
Abstract
:1. Introduction
2. COVID-19 and Gut Microbiota
3. Probiotics as a Useful Treatment in Viral Respiratory Infections
4. NGPs Include Promising Strains
4.1. Faecalibacterium prausnitzii
4.2. Akkermansia muciniphila
4.3. Bacteroides fragilis
4.4. The Emergence of Other Bacteria
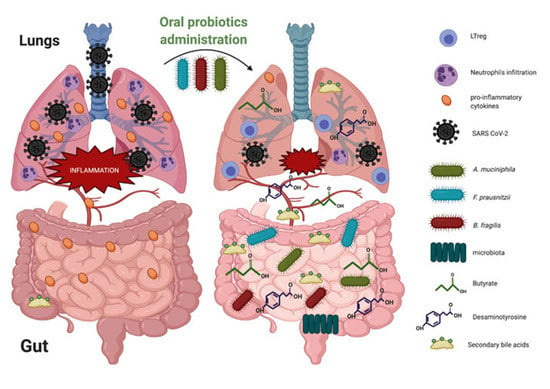
5. Probiotics Administration on Gut–Lung Axis in the Context of Respiratory Tract Infections
6. Probiotic-Derived Metabolites as Beneficial Mediators in Viral Infection?
6.1. Butyrate
6.2. Desaminotyrosine (DAT)
6.3. Secondary Bile Acids
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lai, C.-C.; Shih, T.-P.; Ko, W.-C.; Tang, H.-J.; Hsueh, P.-R. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) and Coronavirus Disease-2019 (COVID-19): The Epidemic and the Challenges. Int. J. Antimicrob. Agents 2020, 55, 105924. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Zhou, L.; Hu, Z.; Zhang, S.; Yang, S.; Tao, Y.; Xie, C.; Ma, K.; Shang, K.; Wang, W.; et al. Dysregulation of Immune Response in Patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Wang, X.; Ni, L.; Di, X.; Ma, B.; Niu, S.; Liu, C.; Reiter, R.J. COVID-19: Melatonin as a Potential Adjuvant Treatment. Life Sci. 2020, 250, 117583. [Google Scholar] [CrossRef] [PubMed]
- Luo, P.; Liu, Y.; Qiu, L.; Liu, X.; Liu, D.; Li, J. Tocilizumab Treatment in COVID-19: A Single Center Experience. J. Med. Virol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Gou, W.; Fu, Y.; Yue, L.; Chen, G.; Cai, X.; Shuai, M.; Xu, F.; Yi, X.; Chen, H.; Zhu, Y.; et al. Gut Microbiota May Underlie the Predisposition of Healthy Individuals to COVID-19. medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Kruglikov, I.L.; Shah, M.; Scherer, P.E. Obesity and Diabetes as Comorbidities for COVID-19: Underlying Mechanisms and the Role of Viral-Bacterial Interactions. eLife 2020, 9. [Google Scholar] [CrossRef]
- Gu, S.; Chen, Y.; Wu, Z.; Chen, Y.; Gao, H.; Lv, L.; Guo, F.; Zhang, X.; Luo, R.; Huang, C.; et al. Alterations of the Gut Microbiota in Patients with COVID-19 or H1N1 Influenza. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Xu, K.; Cai, H.; Shen, Y.; Ni, Q.; Chen, Y.; Hu, S.; Li, J.; Wang, H.; Yu, L.; Huang, H.; et al. Management of corona virus disease-19 (COVID-19): The Zhejiang experience. Zhejiang Da Xue Xue Bao Yi Xue Ban 2020, 49, 147–157. [Google Scholar] [CrossRef]
- Zuo, T.; Liu, Q.; Zhang, F.; Lui, G.C.-Y.; Tso, E.Y.; Yeoh, Y.K.; Chen, Z.; Boon, S.S.; Chan, F.K.; Chan, P.K.; et al. Depicting SARS-CoV-2 Faecal Viral Activity in Association with Gut Microbiota Composition in Patients with COVID-19. Gut 2021, 70, 276–284. [Google Scholar] [CrossRef]
- Dhar, D.; Mohanty, A. Gut Microbiota and Covid-19- Possible Link and Implications. Virus Res. 2020, 285, 198018. [Google Scholar] [CrossRef]
- Zuo, T.; Zhang, F.; Lui, G.C.Y.; Yeoh, Y.K.; Li, A.Y.L.; Zhan, H.; Wan, Y.; Chung, A.C.K.; Cheung, C.P.; Chen, N.; et al. Alterations in Gut Microbiota of Patients With COVID-19 During Time of Hospitalization. Gastroenterology 2020, 159, 944–955. [Google Scholar] [CrossRef]
- Yeoh, Y.K.; Zuo, T.; Lui, G.C.-Y.; Zhang, F.; Liu, Q.; Li, A.Y.; Chung, A.C.; Cheung, C.P.; Tso, E.Y.; Fung, K.S.; et al. Gut Microbiota Composition Reflects Disease Severity and Dysfunctional Immune Responses in Patients with COVID-19. Gut 2021, 70, 698–706. [Google Scholar] [CrossRef]
- Sokol, H.; Contreras, V.; Maisonnasse, P.; Desmons, A.; Delache, B.; Sencio, V.; Machelart, A.; Brisebarre, A.; Humbert, L.; Deryuter, L.; et al. SARS-CoV-2 Infection in Nonhuman Primates Alters the Composition and Functional Activity of the Gut Microbiota. Gut Microbes 2021, 13, 1–19. [Google Scholar] [CrossRef]
- Passioti, M.; Maggina, P.; Megremis, S.; Papadopoulos, N.G. The Common Cold: Potential for Future Prevention or Cure. Curr. Allergy Asthma Rep. 2014, 14, 413. [Google Scholar] [CrossRef]
- Lee, E.-S.; Song, E.-J.; Nam, Y.-D.; Lee, S.-Y. Probiotics in Human Health and Disease: From Nutribiotics to Pharmabiotics. J. Microbiol. 2018, 56, 773–782. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics Consensus Statement on the Scope and Appropriate Use of the Term Probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, Q.; Dong, B.R.; Wu, T. Probiotics for Preventing Acute Upper Respiratory Tract Infections. Cochrane Database Syst. Rev. 2015. [Google Scholar] [CrossRef] [PubMed]
- Vouloumanou, E.K.; Makris, G.C.; Karageorgopoulos, D.E.; Falagas, M.E. Probiotics for the Prevention of Respiratory Tract Infections: A Systematic Review. Int. J. Antimicrob. Agents 2009, 34, 197.e1–197.e10. [Google Scholar] [CrossRef]
- Long, J.D.; Morris, A. Probiotics in Preventing Acute Upper Respiratory Tract Infections. Am. J. Nurs. 2017, 117, 69. [Google Scholar] [CrossRef] [PubMed]
- Weizman, Z.; Asli, G.; Alsheikh, A. Effect of a Probiotic Infant Formula on Infections in Child Care Centers: Comparison of Two Probiotic Agents. Pediatrics 2005, 115, 5–9. [Google Scholar] [CrossRef]
- Belkacem, N.; Serafini, N.; Wheeler, R.; Derrien, M.; Boucinha, L.; Couesnon, A.; Cerf-Bensussan, N.; Gomperts Boneca, I.; Di Santo, J.P.; Taha, M.-K.; et al. Lactobacillus Paracasei Feeding Improves Immune Control of Influenza Infection in Mice. PLoS ONE 2017, 12, e0184976. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.-J.; Lee, Y.-T.; Ngo, V.L.; Cho, Y.-H.; Ko, E.-J.; Hong, S.-M.; Kim, K.-H.; Jang, J.-H.; Oh, J.-S.; Park, M.-K.; et al. Heat-Killed Lactobacillus Casei Confers Broad Protection against Influenza A Virus Primary Infection and Develops Heterosubtypic Immunity against Future Secondary Infection. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz, L.; Delgado, S.; Ruas-Madiedo, P.; Sánchez, B.; Margolles, A. Bifidobacteria and Their Molecular Communication with the Immune System. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, C.B.; Odamaki, T.; Xiao, J. Beneficial Effects of Bifidobacterium Longum Subsp. Longum BB536 on Human Health: Modulation of Gut Microbiome as the Principal Action. J. Funct. Foods 2019, 54, 506–519. [Google Scholar] [CrossRef]
- Mahooti, M.; Abdolalipour, E.; Salehzadeh, A.; Mohebbi, S.R.; Gorji, A.; Ghaemi, A. Immunomodulatory and Prophylactic Effects of Bifidobacterium Bifidum Probiotic Strain on Influenza Infection in Mice. World J. Microbiol. Biotechnol. 2019, 35. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, P.W.; Marchesi, J.R.; Hill, C. Next-Generation Probiotics: The Spectrum from Probiotics to Live Biotherapeutics. Nat. Microbiol. 2017, 2. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-J.; Lin, T.-L.; Tsai, Y.-L.; Wu, T.-R.; Lai, W.-F.; Lu, C.-C.; Lai, H.-C. Next Generation Probiotics in Disease Amelioration. J. Food Drug Anal. 2019, 27, 615–622. [Google Scholar] [CrossRef]
- Lordan, C.; Thapa, D.; Ross, R.P.; Cotter, P.D. Potential for Enriching Next-Generation Health-Promoting Gut Bacteria through Prebiotics and Other Dietary Components. Gut Microbes 2020, 11, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Alameddine, J.; Godefroy, E.; Papargyris, L.; Sarrabayrouse, G.; Tabiasco, J.; Bridonneau, C.; Yazdanbakhsh, K.; Sokol, H.; Altare, F.; Jotereau, F. Faecalibacterium Prausnitzii Skews Human DC to Prime IL10-Producing T Cells Through TLR2/6/JNK Signaling and IL-10, IL-27, CD39, and IDO-1 Induction. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Roychowdhury, S.; Cadnum, J.; Glueck, B.; Obrenovich, M.; Donskey, C.; Cresci, G.A.M. Faecalibacterium Prausnitzii and a Prebiotic Protect Intestinal Health in a Mouse Model of Antibiotic and Clostridium Difficile Exposure. J. Parenter. Enter. Nutr. 2018, 42, 1156–1167. [Google Scholar] [CrossRef]
- Munukka, E.; Rintala, A.; Toivonen, R.; Nylund, M.; Yang, B.; Takanen, A.; Hänninen, A.; Vuopio, J.; Huovinen, P.; Jalkanen, S.; et al. Faecalibacterium Prausnitzii Treatment Improves Hepatic Health and Reduces Adipose Tissue Inflammation in High-Fat Fed Mice. ISME J. 2017, 11, 1667–1679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savin, K.W.; Zawadzki, J.; Auldist, M.J.; Wang, J.; Ram, D.; Rochfort, S.; Cocks, B.G. Faecalibacterium Diversity in Dairy Cow Milk. PLoS ONE 2019, 14, e0221055. [Google Scholar] [CrossRef] [Green Version]
- Fujimura, K.E.; Sitarik, A.R.; Havstad, S.; Lin, D.L.; Levan, S.; Fadrosh, D.; Panzer, A.R.; LaMere, B.; Rackaityte, E.; Lukacs, N.W.; et al. Neonatal Gut Microbiota Associates with Childhood Multisensitized Atopy and T Cell Differentiation. Nat. Med. 2016, 22, 1187–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, J.; Zhang, L.; Lin, W.; Tang, W.; Chan, F.K.L.; Ng, S.C. Review Article: Probiotics, Prebiotics and Dietary Approaches during COVID-19 Pandemic. Trends Food Sci. Technol. 2021, 108, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Farias, C.; Slezak, K.; Fuller, Z.; Duncan, A.; Holtrop, G.; Louis, P. Effect of Inulin on the Human Gut Microbiota: Stimulation of Bifidobacterium Adolescentis and Faecalibacterium Prausnitzii. Br. J. Nutr. 2009, 101, 541–550. [Google Scholar] [CrossRef] [Green Version]
- Nogacka, A.M.; Salazar, N.; Arboleya, S.; Ruas-Madiedo, P.; Mancabelli, L.; Suarez, A.; Martinez-Faedo, C.; Ventura, M.; Tochio, T.; Hirano, K.; et al. In Vitro Evaluation of Different Prebiotics on the Modulation of Gut Microbiota Composition and Function in Morbid Obese and Normal-Weight Subjects. Int. J. Mol. Sci. 2020, 21, 906. [Google Scholar] [CrossRef] [Green Version]
- Finegold, S.M.; Li, Z.; Summanen, P.H.; Downes, J.; Thames, G.; Corbett, K.; Dowd, S.; Krak, M.; Heber, D. Xylooligosaccharide Increases Bifidobacteria but Not Lactobacilli in Human Gut Microbiota. Food Funct. 2014, 5, 436–445. [Google Scholar] [CrossRef] [PubMed]
- Lensu, S.; Pariyani, R.; Mäkinen, E.; Yang, B.; Saleem, W.; Munukka, E.; Lehti, M.; Driuchina, A.; Lindén, J.; Tiirola, M.; et al. Prebiotic Xylo-Oligosaccharides Ameliorate High-Fat-Diet-Induced Hepatic Steatosis in Rats. Nutrients 2020, 12, 3225. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Li, Q.; Cheng, L.; Buch, H.; Zhang, F. Akkermansia muciniphila Is a Promising Probiotic. Microb. Biotechnol. 2019, 12, 1109–1125. [Google Scholar] [CrossRef] [Green Version]
- Ansaldo, E.; Slayden, L.C.; Ching, K.L.; Koch, M.A.; Wolf, N.K.; Plichta, D.R.; Brown, E.M.; Graham, D.B.; Xavier, R.J.; Moon, J.J.; et al. Akkermansia muciniphila Induces Intestinal Adaptive Immune Responses during Homeostasis. Science 2019, 364, 1179–1184. [Google Scholar] [CrossRef]
- Bian, X.; Wu, W.; Yang, L.; Lv, L.; Wang, Q.; Li, Y.; Ye, J.; Fang, D.; Wu, J.; Jiang, X.; et al. Administration of Akkermansia muciniphila Ameliorates Dextran Sulfate Sodium-Induced Ulcerative Colitis in Mice. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Ashrafian, F.; Shahriary, A.; Behrouzi, A.; Moradi, H.R.; Keshavarz Azizi Raftar, S.; Lari, A.; Hadifar, S.; Yaghoubfar, R.; Ahmadi Badi, S.; Khatami, S.; et al. Akkermansia muciniphila-Derived Extracellular Vesicles as a Mucosal Delivery Vector for Amelioration of Obesity in Mice. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Zhu, L.; Lu, X.; Liu, L.; Voglmeir, J.; Zhong, X.; Yu, Q. Akkermansia muciniphila Protects Intestinal Mucosa from Damage Caused by S. Pullorum by Initiating Proliferation of Intestinal Epithelium. Vet. Res. 2020, 51. [Google Scholar] [CrossRef] [Green Version]
- Quintana-Hayashi, M.; Padra, M.; Padra, J.; Benktander, J.; Lindén, S. Mucus-Pathogen Interactions in the Gastrointestinal Tract of Farmed Animals. Microorganisms 2018, 6, 55. [Google Scholar] [CrossRef] [Green Version]
- Ridley, C.; Thornton, D.J. Mucins: The Frontline Defence of the Lung. Biochem. Soc. Trans. 2018, 46, 1099–1106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ehre, C.; Worthington, E.N.; Liesman, R.M.; Grubb, B.R.; Barbier, D.; O’Neal, W.K.; Sallenave, J.-M.; Pickles, R.J.; Boucher, R.C. Overexpressing Mouse Model Demonstrates the Protective Role of Muc5ac in the Lungs. Proc. Natl. Acad. Sci. USA 2012, 109, 16528–16533. [Google Scholar] [CrossRef] [Green Version]
- Ottman, N.; Geerlings, S.Y.; Aalvink, S.; de Vos, W.M.; Belzer, C. Action and Function of Akkermansia muciniphila in Microbiome Ecology, Health and Disease. Best Pract. Res. Clin. Gastroenterol. 2017, 31, 637–642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plovier, H.; Everard, A.; Druart, C.; Depommier, C.; Van Hul, M.; Geurts, L.; Chilloux, J.; Ottman, N.; Duparc, T.; Lichtenstein, L.; et al. A Purified Membrane Protein from Akkermansia muciniphila or the Pasteurized Bacterium Improves Metabolism in Obese and Diabetic Mice. Nat. Med. 2017, 23, 107–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Xiang, R.; Wang, R.; Zhang, B.; Gong, W.; Zhang, J.; Zhang, M.; Wang, M. The Variable Oligomeric State of Amuc_1100 from Akkermansia muciniphila. J. Struct. Biol. 2020, 212, 107593. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Zhang, Q.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. A Potential Species of Next-Generation Probiotics? The Dark and Light Sides of Bacteroides Fragilis in Health. Food Res. Int. 2019, 126, 108590. [Google Scholar] [CrossRef]
- Zhang, W.; Zhu, B.; Xu, J.; Liu, Y.; Qiu, E.; Li, Z.; Li, Z.; He, Y.; Zhou, H.; Bai, Y.; et al. Bacteroides Fragilis Protects against Antibiotic-Associated Diarrhea in Rats by Modulating Intestinal Defenses. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Casterline, B.W.; Hecht, A.L.; Choi, V.M.; Bubeck Wardenburg, J. The Bacteroides Fragilis Pathogenicity Island Links Virulence and Strain Competition. Gut Microbes 2017, 8, 374–383. [Google Scholar] [CrossRef] [Green Version]
- Round, J.L.; Mazmanian, S.K. Inducible Foxp3+ Regulatory T-Cell Development by a Commensal Bacterium of the Intestinal Microbiota. Proc. Natl. Acad. Sci. USA 2010, 107, 12204–12209. [Google Scholar] [CrossRef] [Green Version]
- Ramakrishna, C.; Kujawski, M.; Chu, H.; Li, L.; Mazmanian, S.K.; Cantin, E.M. Bacteroides Fragilis Polysaccharide A Induces IL-10 Secreting B and T Cells That Prevent Viral Encephalitis. Nat. Commun. 2019, 10, 2153. [Google Scholar] [CrossRef] [Green Version]
- Alvarez, C.A.; Jones, M.B.; Hambor, J.; Cobb, B.A. Characterization of Polysaccharide A Response Reveals Interferon Responsive Gene Signature and Immunomodulatory Marker Expression. Front. Immunol. 2020, 11, 556813. [Google Scholar] [CrossRef]
- Zhao, X.; Yang, J.; Ju, Z.; Wu, J.; Wang, L.; Lin, H.; Sun, S. Clostridium Butyricum Ameliorates Salmonella Enteritis Induced Inflammation by Enhancing and Improving Immunity of the Intestinal Epithelial Barrier at the Intestinal Mucosal Level. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef] [Green Version]
- Tomaro-Duchesneau, C.; LeValley, S.L.; Roeth, D.; Sun, L.; Horrigan, F.T.; Kalkum, M.; Hyser, J.M.; Britton, R.A. Discovery of a Bacterial Peptide as a Modulator of GLP-1 and Metabolic Disease. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef] [Green Version]
- Ma, S.; Yeom, J.; Lim, Y.-H. Dairy Propionibacterium Freudenreichii Ameliorates Acute Colitis by Stimulating MUC2 Expression in Intestinal Goblet Cell in a DSS-Induced Colitis Rat Model. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Kanmani, P.; Clua, P.; Vizoso-Pinto, M.G.; Rodriguez, C.; Alvarez, S.; Melnikov, V.; Takahashi, H.; Kitazawa, H.; Villena, J. Respiratory Commensal Bacteria Corynebacterium Pseudodiphtheriticum Improves Resistance of Infant Mice to Respiratory Syncytial Virus and Streptococcus Pneumoniae Superinfection. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Li, S.; Wang, N.; Tan, H.-Y.; Zhang, Z.; Feng, Y. The Cross-Talk Between Gut Microbiota and Lungs in Common Lung Diseases. Front. Microbiol. 2020, 11, 301. [Google Scholar] [CrossRef]
- Vaughan, A.; Frazer, Z.A.; Hansbro, P.M.; Yang, I.A. COPD and the Gut-Lung Axis: The Therapeutic Potential of Fibre. J. Thorac. Dis. 2019, 11, S2173–S2180. [Google Scholar] [CrossRef]
- Antosca, K.M.; Chernikova, D.A.; Price, C.E.; Ruoff, K.L.; Li, K.; Guill, M.F.; Sontag, N.R.; Morrison, H.G.; Hao, S.; Drumm, M.L.; et al. Altered Stool Microbiota of Infants with Cystic Fibrosis Shows a Reduction in Genera Associated with Immune Programming from Birth. J. Bacteriol. 2019, 201. [Google Scholar] [CrossRef] [Green Version]
- Bradley, C.P.; Teng, F.; Felix, K.M.; Sano, T.; Naskar, D.; Block, K.E.; Huang, H.; Knox, K.S.; Littman, D.R.; Wu, H.-J.J. Segmented Filamentous Bacteria Provoke Lung Autoimmunity by Inducing Gut-Lung Axis Th17 Cells Expressing Dual TCRs. Cell Host Microbe 2017, 22, 697–704.e4. [Google Scholar] [CrossRef]
- Hufnagl, K.; Pali-Schöll, I.; Roth-Walter, F.; Jensen-Jarolim, E. Dysbiosis of the Gut and Lung Microbiome Has a Role in Asthma. Semin. Immunopathol. 2020, 42, 75–93. [Google Scholar] [CrossRef] [Green Version]
- Hanada, S.; Pirzadeh, M.; Carver, K.Y.; Deng, J.C. Respiratory Viral Infection-Induced Microbiome Alterations and Secondary Bacterial Pneumonia. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Yildiz, S.; Mazel-Sanchez, B.; Kandasamy, M.; Manicassamy, B.; Schmolke, M. Influenza A Virus Infection Impacts Systemic Microbiota Dynamics and Causes Quantitative Enteric Dysbiosis. Microbiome 2018, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, N.; Zheng, B.; Yao, J.; Guo, L.; Zuo, J.; Wu, L.; Zhou, J.; Liu, L.; Guo, J.; Ni, S.; et al. Influence of H7N9 Virus Infection and Associated Treatment on Human Gut Microbiota. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef]
- Wang, B.; Yao, M.; Lv, L.; Ling, Z.; Li, L. The Human Microbiota in Health and Disease. Engineering 2017, 3, 71–82. [Google Scholar] [CrossRef]
- Yu, B.; Dai, C.; Chen, J.; Deng, L.; Wu, X.; Wu, S.; Zhao, C.; Jiang, Z.; Chen, X. Dysbiosis of Gut Microbiota Induced the Disorder of Helper T Cells in Influenza Virus-Infected Mice. Hum. Vaccines Immunother. 2015, 11, 1140–1146. [Google Scholar] [CrossRef] [Green Version]
- Ichinohe, T.; Pang, I.K.; Kumamoto, Y.; Peaper, D.R.; Ho, J.H.; Murray, T.S.; Iwasaki, A. Microbiota Regulates Immune Defense against Respiratory Tract Influenza A Virus Infection. Proc. Natl. Acad. Sci. USA 2011, 108, 5354–5359. [Google Scholar] [CrossRef] [Green Version]
- Dumas, A.; Bernard, L.; Poquet, Y.; Lugo-Villarino, G.; Neyrolles, O. The Role of the Lung Microbiota and the Gut-Lung Axis in Respiratory Infectious Diseases. Cell. Microbiol. 2018, 20, e12966. [Google Scholar] [CrossRef] [Green Version]
- Anand, S.; Mande, S.S. Diet, Microbiota and Gut-Lung Connection. Front. Microbiol. 2018, 9, 2147. [Google Scholar] [CrossRef] [PubMed]
- Postler, T.S.; Ghosh, S. Understanding the Holobiont: How Microbial Metabolites Affect Human Health and Shape the Immune System. Cell Metab. 2017, 26, 110–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sittipo, P.; Shim, J.; Lee, Y. Microbial Metabolites Determine Host Health and the Status of Some Diseases. Int. J. Mol. Sci. 2019, 20, 5296. [Google Scholar] [CrossRef] [Green Version]
- Haak, B.W.; Littmann, E.R.; Chaubard, J.-L.; Pickard, A.J.; Fontana, E.; Adhi, F.; Gyaltshen, Y.; Ling, L.; Morjaria, S.M.; Peled, J.U.; et al. Impact of Gut Colonization with Butyrate Producing Microbiota on Respiratory Viral Infection Following Allo-HCT. Blood 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steed, A.L.; Christophi, G.P.; Kaiko, G.E.; Sun, L.; Goodwin, V.M.; Jain, U.; Esaulova, E.; Artyomov, M.N.; Morales, D.J.; Holtzman, M.J.; et al. The Microbial Metabolite Desaminotyrosine Protects from Influenza through Type I Interferon. Science 2017, 357, 498–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, L.; Han, W.; Du, J.; Yang, X.; Duan, M.; Xu, C.; Zeng, Z.; Chen, W.; Chen, J. Chenodeoxycholic Acid from Bile Inhibits Influenza A Virus Replication via Blocking Nuclear Export of Viral Ribonucleoprotein Complexes. Molecules 2018, 23, 3315. [Google Scholar] [CrossRef] [Green Version]
- LeBlanc, J.G.; Milani, C.; de Giori, G.S.; Sesma, F.; van Sinderen, D.; Ventura, M. Bacteria as Vitamin Suppliers to Their Host: A Gut Microbiota Perspective. Curr. Opin. Biotechnol. 2013, 24, 160–168. [Google Scholar] [CrossRef]
- Konopelski, P.; Ufnal, M. Indoles—Gut Bacteria Metabolites of Tryptophan with Pharmacotherapeutic Potential. Curr. Drug Metab. 2018, 19, 883–890. [Google Scholar] [CrossRef]
- Xie, R.; Jiang, P.; Lin, L.; Jiang, J.; Yu, B.; Rao, J.; Liu, H.; Wei, W.; Qiao, Y. Oral Treatment with Lactobacillus Reuteri Attenuates Depressive-like Behaviors and Serotonin Metabolism Alterations Induced by Chronic Social Defeat Stress. J. Psychiatr. Res. 2020, 122, 70–78. [Google Scholar] [CrossRef]
- Bravo, J.A.; Forsythe, P.; Chew, M.V.; Escaravage, E.; Savignac, H.M.; Dinan, T.G.; Bienenstock, J.; Cryan, J.F. Ingestion of Lactobacillus Strain Regulates Emotional Behavior and Central GABA Receptor Expression in a Mouse via the Vagus Nerve. Proc. Natl. Acad. Sci. USA 2011, 108, 16050–16055. [Google Scholar] [CrossRef] [Green Version]
- Levy, M.; Thaiss, C.A.; Elinav, E. Metabolites: Messengers between the Microbiota and the Immune System. Genes Dev. 2016, 30, 1589–1597. [Google Scholar] [CrossRef]
- Breyner, N.M.; Michon, C.; de Sousa, C.S.; Vilas Boas, P.B.; Chain, F.; Azevedo, V.A.; Langella, P.; Chatel, J.M. Microbial Anti-Inflammatory Molecule (MAM) from Faecalibacterium Prausnitzii Shows a Protective Effect on DNBS and DSS-Induced Colitis Model in Mice through Inhibition of NF-ΚB Pathway. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Cani, P.D.; de Vos, W.M. Next-Generation Beneficial Microbes: The Case of Akkermansia muciniphila. Front. Microbiol. 2017, 8, 1765. [Google Scholar] [CrossRef]
- Ottman, N.; Reunanen, J.; Meijerink, M.; Pietilä, T.E.; Kainulainen, V.; Klievink, J.; Huuskonen, L.; Aalvink, S.; Skurnik, M.; Boeren, S.; et al. Pili-like Proteins of Akkermansia muciniphila Modulate Host Immune Responses and Gut Barrier Function. PLoS ONE 2017, 12, e0173004. [Google Scholar] [CrossRef]
- Fernández, J.; Redondo-Blanco, S.; Gutiérrez-del-Río, I.; Miguélez, E.M.; Villar, C.J.; Lombó, F. Colon Microbiota Fermentation of Dietary Prebiotics towards Short-Chain Fatty Acids and Their Roles as Anti-Inflammatory and Antitumour Agents: A Review. J. Funct. Foods 2016, 25, 511–522. [Google Scholar] [CrossRef]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, L.; Kelly, C.J.; Battista, K.D.; Schaefer, R.; Lanis, J.M.; Alexeev, E.E.; Wang, R.X.; Onyiah, J.C.; Kominsky, D.J.; Colgan, S.P. Microbial-Derived Butyrate Promotes Epithelial Barrier Function through IL-10 Receptor–Dependent Repression of Claudin-2. J. Immunol. 2017, 199, 2976–2984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakraborty, K.; Raundhal, M.; Chen, B.B.; Morse, C.; Tyurina, Y.Y.; Khare, A.; Oriss, T.B.; Huff, R.; Lee, J.S.; St Croix, C.M.; et al. The Mito-DAMP Cardiolipin Blocks IL-10 Production Causing Persistent Inflammation during Bacterial Pneumonia. Nat. Commun. 2017, 8, 13944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dang, A.T.; Marsland, B.J. Microbes, Metabolites, and the Gut–Lung Axis. Mucosal Immunol. 2019, 12, 843–850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murota, K.; Nakamura, Y.; Uehara, M. Flavonoid Metabolism: The Interaction of Metabolites and Gut Microbiota. Biosci. Biotechnol. Biochem. 2018, 82, 600–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An Overview. J. Nutr. Sci. 2016, 5. [Google Scholar] [CrossRef] [Green Version]
- Lobel, L.; Garrett, W.S. Take DAT, Flu! Immunity 2017, 47, 400–402. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Chang, K.-O. Inhibitory Effects of Bile Acids and Synthetic Farnesoid X Receptor Agonists on Rotavirus Replication. J. Virol. 2011, 85, 12570–12577. [Google Scholar] [CrossRef] [Green Version]
- Guo, C.; Xie, S.; Chi, Z.; Zhang, J.; Liu, Y.; Zhang, L.; Zheng, M.; Zhang, X.; Xia, D.; Ke, Y.; et al. Bile Acids Control Inflammation and Metabolic Disorder through Inhibition of NLRP3 Inflammasome. Immunity 2016, 45, 944. [Google Scholar] [CrossRef] [Green Version]
- Hang, S.; Paik, D.; Yao, L.; Kim, E.; Trinath, J.; Lu, J.; Ha, S.; Nelson, B.N.; Kelly, S.P.; Wu, L.; et al. Bile Acid Metabolites Control TH17 and Treg Cell Differentiation. Nature 2019, 576, 143–148. [Google Scholar] [CrossRef]
- Nagpal, R.; Mainali, R.; Ahmadi, S.; Wang, S.; Singh, R.; Kavanagh, K.; Kitzman, D.W.; Kushugulova, A.; Marotta, F.; Yadav, H. Gut Microbiome and Aging: Physiological and Mechanistic Insights. Nutr. Healthy Aging 2018, 4, 267–285. [Google Scholar] [CrossRef] [Green Version]
- Goyal, P.; Choi, J.J.; Pinheiro, L.C.; Schenck, E.J.; Chen, R.; Jabri, A.; Satlin, M.J.; Campion, T.R.; Nahid, M.; Ringel, J.B.; et al. Clinical Characteristics of Covid-19 in New York City. N. Engl. J. Med. 2020, 382, 2372–2374. [Google Scholar] [CrossRef]
- Lake, M.A. What We Know so Far: COVID-19 Current Clinical Knowledge and Research. Clin. Med. 2020, 20, 124–127. [Google Scholar] [CrossRef] [Green Version]


| Metabolites | Main Bacteria Responsible for Production | Respiratory Tract Infection or Other Pathology | Mode of Action | Reference |
|---|---|---|---|---|
| Short-chain fatty acids (SCFAs) Butyrate | Faecalibacterium | Lower respiratory tract infection Influenza virus | Regulate inflammation, promote monocytes, decrease neutrophils, maintain gut barrier function. | [76] |
| Desaminotyrosine (DAT) | Clostridium orbiscindens | Influenza virus H1N1 | Inhibit virus replication (interferons), decrease lung immunopathology. | [77] |
| Secondary bile acid | Bacteroides | Influenza virus H5N1 | Inhibit virus replication, has anti-inflammatory properties. | [78] |
| Folate | Lactobacillus, Bifidobacterium | Gastric colorectal cancer | Involved in many metabolic pathways. | [79] |
| Indole derivatives | Bacteroides thetaiotaomicron | Metabolic syndrome, Psychiatric diseases | Affect host intestinal inflammation. | [80] |
| Gut-derived Neuro-transmitters (Serotonin, GABA, Histamine) | L. reuteri Lactobacillus, BifidobacteriumL. reuteri | Depression Anxiety and depression Allergy | Regulate numerous physiological processes, inhibit neurotransmitter (GABA), have several roles in immune functions. | [81] [82] [83] |
| Microbial Anti-Inflammatory molecules (MAMs) | F. prausnitzii | Colitis | Have anti-inflammatory properties | [84] |
| Amuc_1100 | A. muciniphila | Obesity | Regulate host immunological homeostasis, and improve of gut barrier function | [85,86] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gautier, T.; David-Le Gall, S.; Sweidan, A.; Tamanai-Shacoori, Z.; Jolivet-Gougeon, A.; Loréal, O.; Bousarghin, L. Next-Generation Probiotics and Their Metabolites in COVID-19. Microorganisms 2021, 9, 941. https://doi.org/10.3390/microorganisms9050941
Gautier T, David-Le Gall S, Sweidan A, Tamanai-Shacoori Z, Jolivet-Gougeon A, Loréal O, Bousarghin L. Next-Generation Probiotics and Their Metabolites in COVID-19. Microorganisms. 2021; 9(5):941. https://doi.org/10.3390/microorganisms9050941
Chicago/Turabian StyleGautier, Thomas, Sandrine David-Le Gall, Alaa Sweidan, Zohreh Tamanai-Shacoori, Anne Jolivet-Gougeon, Olivier Loréal, and Latifa Bousarghin. 2021. "Next-Generation Probiotics and Their Metabolites in COVID-19" Microorganisms 9, no. 5: 941. https://doi.org/10.3390/microorganisms9050941