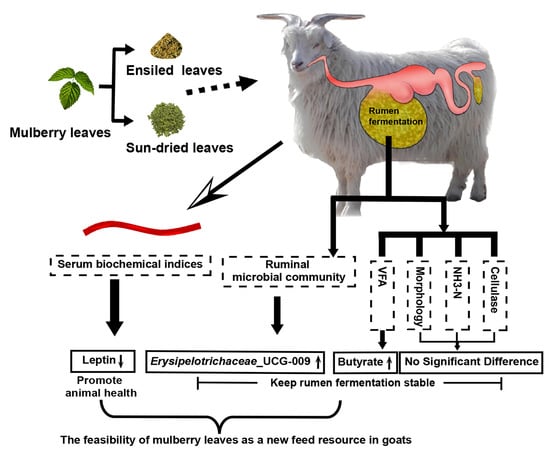
Comparison of Rumen Microbiota and Serum Biochemical Indices in White Cashmere Goats Fed Ensiled or Sun-Dried Mulberry Leaves
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Diet, Animal Management, and Sample Collection
2.3. DNA Extraction and 16S rRNA Gene Sequencing
2.4. Sequencing Analyses
2.5. Bacterial Function Prediction
2.6. qPCR Analysis
2.7. Volatile Fatty Acid, NH3-N and Carboxymethylcellulase Activity Assays
2.8. Rumen Morphological Examination
2.9. Serum Biochemical Analysis
2.10. Statistical Analysis
3. Results
3.1. Sequences Across Different Dietary Groups
3.2. Analysis of Similarity
3.3. Effects of Different Feed Types on the Composition of Ruminal Bacteria
3.4. Functional Predictions of Rumen Bacteria
3.5. Proportion of Selected Microbiota in the Rumen
3.6. Rumen Morphological and Functional Parameters
3.7. Biochemical Analysis of Serum Samples
3.8. Correlation Analysis
4. Discussion
4.1. Changes in Rumen Fermentation Parameters and Serum Biochemical Indicators
4.2. Comparison of the Composition of Ruminal Microbiota after Adding Mulberry Leaves to the Diets
4.3. Relationships among Rumen Microbiota, Fermentation Parameters and Serum Biochemical Indices
4.4. Predicted Functions of Ruminal Bacteria in Goats
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Li, R.; Chen, D.; Wang, T.; Wan, Y.; Li, R.; Fang, R.; Wang, Y.; Hu, F.; Zhou, H.; Li, L.; et al. High throughput deep degradome sequencing reveals microRNAs and their targets in response to drought stress in mulberry (Morus alba). PLoS ONE 2017, 12, e172883. [Google Scholar] [CrossRef]
- Zhou, Z.; Zhou, B.; Ren, L.; Meng, Q. Effect of ensiled mulberry leaves and sun-dried mulberry fruit pomace on finishing steer growth performance, blood biochemical parameters, and carcass characteristics. PLoS ONE 2014, 9, e85406. [Google Scholar] [CrossRef]
- CFIC. Tables of feed composition and nutritive in China (2019 thirty edition). China Feed 2019, 22, 111–116. [Google Scholar] [CrossRef]
- Ouyang, J.; Wang, M.; Hou, Q.; Feng, D.; Pi, Y.; Zhao, W. Effects of dietary mulberry leaf powder in concentrate on the rumen fermentation and ruminal epithelium in fattening Hu sheep. Animals 2019, 9, 218. [Google Scholar] [CrossRef] [Green Version]
- Min, H.K.; Kapoor, A.; Fuchs, M.; Mirshahi, F.; Zhou, H.; Maher, J.; Kellum, J.; Warnick, R.; Contos, M.J.; Sanyal, A.J. Increased hepatic synthesis and dysregulation of cholesterol metabolism is associated with the severity of nonalcoholic fatty liver disease. Cell Metab. 2012, 15, 665–674. [Google Scholar] [CrossRef] [Green Version]
- Al-kirshi, R.; Alimon, A.R.; Zulkifli, I.; Sazili, A.; Zahari, M.W.; Ivan, M. Utilization of mulberry leaf meal (Morus alba) as protein supplement in diets for laying hens. Ital. J. Anim. Sci. 2010, 9, e51. [Google Scholar] [CrossRef] [Green Version]
- Chi, C.V.; Verstegen, M.W.A.; Hendriks, W.H.; Pham, K.T. The nutritive value of mulberry leaves (Morus alba) and partial replacement of cotton seed in rations on the performance of growing Vietnamese cattle. Asian Aust. J. Anim. Sci. 2011, 24, 1233–1242. [Google Scholar]
- Kandylis, K.; Hadjigeorgiou, I.; Harizanis, P. The nutritive value of mulberry leaves (Morus alba) as a feed supplement for sheep. Trop. Anim. Health Prod. 2009, 41, 17–24. [Google Scholar] [CrossRef]
- Saad-Allah, K.M.; Youssef, M.S. Phytochemical and genetic characterization of five quinoa (Chenopodium quinoa Willd.) genotypes introduced to Egypt. Physiol. Mol. Biol. Plants 2018, 24, 617–629. [Google Scholar] [CrossRef]
- Zardo, I.; Rodrigues, N.P.; Sarkis, J.R.; Marczak, L.D. Extraction and identification by mass spectrometry of phenolic compounds from canola seed cake. J. Sci. Food Agr. 2020, 100, 578–586. [Google Scholar] [CrossRef]
- Getachew, G.; Pittroff, W.; Putnam, D.H.; Dandekar, A.; Goyal, S.; Depeters, E.J. The influence of addition of gallic acid, tannic acid, or quebracho tannins to alfalfa hay on in vitro rumen fermentation and microbial protein synthesis. Anim. Feed Sci. Tech. 2008, 140, 444–461. [Google Scholar] [CrossRef]
- Sánchez, M.D. Mulberry: An exceptional forage available almost worldwide. In Mulberry for Animal Production; Sánchez, M.D., Ed.; Food and agriculture organization (FAO) of United Nations: Rome, Italy, 2002; pp. 189–271. [Google Scholar]
- Askar, A.R.; Nassar, M.S.; Badawy, H.S.; Eid, E.Y.; Guada, J.A.; Farid, M.F.A. Recovered energy and efficiency of digestion in sheep and goats fed Atriplex nummularia compared to alfalfa hay. Livest. Sci. 2016, 194, 1–6. [Google Scholar] [CrossRef]
- He, J.; Feng, Y.; Ouyang, H.; Yu, B.; Chang, Y.; Pan, G.; Dong, G.; Wang, T.; Gao, X. A sensitive LC–MS/MS method for simultaneous determination of six flavonoids in rat plasma: Application to a pharmacokinetic study of total flavonoids from mulberry leaves. J. Pharm. Biomed. Anal. 2013, 84, 189–195. [Google Scholar] [CrossRef]
- Yuan, Q.; Xie, Y.; Wang, W.; Yan, Y.; Ye, H.; Jabbar, S.; Zeng, X. Extraction optimization, characterization and antioxidant activity in vitro of polysaccharides from mulberry (Morus alba L.) leaves. Carbohyd. Polym. 2015, 128, 52–62. [Google Scholar] [CrossRef]
- Sugiyama, M.; Katsube, T.; Koyama, A.; Itamura, H. Effect of solar radiation on the functional components of mulberry (Morus alba L.) leaves. J. Sci. Food Agr. 2016, 96, 3915–3921. [Google Scholar] [CrossRef]
- Prajapati, V.S.; Purohit, H.J.; Raje, D.V.; Parmar, N.; Patel, A.B.; Jones, O.A.H.; Joshi, C.G. The effect of a high-roughage diet on the metabolism of aromatic compounds by rumen microbes: A metagenomic study using Mehsani buffalo (Bubalus bubalis). Appl. Microbiol. Biot. 2016, 100, 1319–1331. [Google Scholar] [CrossRef]
- Sun, Z.; Yu, Z.; Wang, B. Perilla frutescens leaf alters the rumen microbial community of lactating dairy cows. Microorganisms 2019, 7, 562. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Li, C.; Li, F.; Wang, X.; Zhang, X.; Liu, T.; Nian, F.; Yue, X.; Li, F.; Pan, X.; et al. Effects of early feeding on the host rumen transcriptome and bacterial diversity in lambs. Sci. Rep. 2016, 6, 32479. [Google Scholar] [CrossRef]
- Faubladier, C.; Julliand, V.; Danel, J.; Philippeau, C. Bacterial carbohydrate-degrading capacity in foal faeces: Changes from birth to pre-weaning and the impact of maternal supplementation with fermented feed products. Br. J. Nutr. 2013, 110, 1040–1052. [Google Scholar] [CrossRef] [Green Version]
- Ding, S.; Fang, J.; Liu, G.; Veeramuthu, D.; Abdullah, A.N.; Yin, Y. The impact of different levels of cysteine on the plasma metabolomics and intestinal microflora of sows from late pregnancy to lactation. Food Funct. 2019, 10, 691–702. [Google Scholar] [CrossRef]
- Niu, Y.; Meng, Q.; Li, S.; Ren, L.; Zhou, B.; Schonewille, T.; Zhou, Z. Effects of diets supplemented with ensiled mulberry leaves and sun-dried mulberry fruit pomace on the ruminal bacterial and archaeal community composition of finishing steers. PLoS ONE 2016, 11, e156836. [Google Scholar] [CrossRef] [PubMed]
- Huyen, N.T.; Wanapat, M.; Navanukraw, C. Effect of mulberry leaf pellet (MUP) supplementation on rumen fermentation and nutrient digestibility in beef cattle fed on rice straw-based diets. Anim. Feed Sci. Tech. 2012, 175, 8–15. [Google Scholar] [CrossRef]
- Shao, Y.; Zhou, J.; Xiong, X.; Zou, L.; Kong, X.; Tan, B.; Yin, Y. Differences in gut microbial and serum biochemical indices between sows with different productive capacities during perinatal period. Front. Microbiol. 2020, 10, 3047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Liu, Y.; Wan, X.; Huang, Z.; Liu, B.; Zhao, C. Regulatory efficacy of the polyunsaturated fatty acids from microalgae Spirulina platensis on lipid metabolism and gut microbiota in high-fat diet rats. Int. J. Mol. Sci. 2018, 19, 3075. [Google Scholar] [CrossRef] [Green Version]
- He, J.; He, Y.; Pan, D.; Cao, J.; Sun, Y.; Zeng, X. Associations of gut microbiota with heat stress-induced changes of growth, fat deposition, intestinal morphology, and antioxidant capacity in ducks. Front. Microbiol. 2019, 10, 903. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [Green Version]
- Koetschan, C.; Kittelmann, S.; Lu, J.; Al-Halbouni, D.; Jarvis, G.N.; Muller, T.; Wolf, M.; Janssen, P.H. Internal transcribed spacer 1 secondary structure analysis reveals a common core throughout the anaerobic fungi (Neocallimastigomycota). PLoS ONE 2014, 9, e91928. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Bittinger, K.; Bushman, F.D.; DeSantis, T.Z.; Andersen, G.L.; Knight, R. PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics 2010, 26, 266–267. [Google Scholar] [CrossRef] [Green Version]
- Amato, K.R.; Yeoman, C.J.; Kent, A.; Righini, N.; Carbonero, F.; Estrada, A.; Gaskins, H.R.; Stumpf, R.M.; Yildirim, S.; Torralba, M.; et al. Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J. 2013, 7, 1344–1353. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [Green Version]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [Green Version]
- Jami, E.; Israel, A.; Kotser, A.; Mizrahi, I. Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 2013, 7, 1069–1079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Jiang, B.; Liu, J.; Zhao, X.; Liu, Q.; Hu, X. Genome sequence analysis of a flocculant-producing bacterium. Paenibacillus Shenyangensis Biotechnol. Lett. 2016, 38, 447–453. [Google Scholar] [CrossRef]
- Wang, Y.; Cao, P.; Wang, L.; Zhao, Z.; Chen, Y.; Yang, Y. Bacterial community diversity associated with different levels of dietary nutrition in the rumen of sheep. Appl. Microbiol. Biot. 2017, 101, 3717–3728. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.; Li, X.; Beauchemin, K.A.; Tan, Z.; Tang, S.; Zhou, C. Rumen development process in goats as affected by supplemental feeding v. grazing: Age-related anatomic development, functional achievement and microbial colonisation. Brit. J. Nutr. 2015, 113, 888–900. [Google Scholar] [CrossRef] [Green Version]
- Broderick, G.A.; Kang, J.H. Automated simultaneous determination of ammonia and total amino acids in ruminal fluid and in vitro media. J. Dairy Sci. 1980, 63, 64–75. [Google Scholar] [CrossRef]
- Li, D.; Zhang, Y.; Cui, Z.; He, L.; Chen, W.; Meng, Q.; Ren, L. Effects of phytoecdysteroids (PEDS) extracted from Cyanotis arachnoidea on rumen fermentation, enzyme activity and microbial efficiency in a continuous-culture system. PLoS ONE 2016, 11, e153584. [Google Scholar] [CrossRef]
- Barter, P.J. Lipoprotein metabolism and CKD: Overview. Clin. Exp. Nephrol. 2014, 18, 243–246. [Google Scholar] [CrossRef]
- Abourjaili, G.; Shtaynberg, N.; Wetz, R.V.; Costantino, T.; Abela, G.S. Current concepts in triglyceride metabolism, pathophysiology, and treatment. Metabolism 2010, 59, 1210–1220. [Google Scholar] [CrossRef] [PubMed]
- Banks, W.A.; Farr, S.A.; Salameh, T.S.; Niehoff, M.L.; Rhea, E.M.; Morley, J.E.; Hanson, A.J.; Hansen, K.M.; Craft, S. Triglycerides cross the blood–brain barrier and induce central leptin and insulin receptor resistance. Int. J. Obes. 2018, 42, 391–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiao, J.; Huang, J.; Zhou, C.; Tan, Z. Taxonomic identification of ruminal epithelial bacterial diversity during rumen development in goats. Appl. Environ. Microb. 2015, 81, 3502–3509. [Google Scholar] [CrossRef] [Green Version]
- Galbraith, M.L.; Vorachek, W.R.; Estill, C.T.; Whanger, P.D.; Bobe, G.; Davis, T.Z.; Hall, J.A. Rumen microorganisms decrease bioavailability of inorganic selenium supplements. Biol. Trace Elem. Res. 2016, 171, 338–343. [Google Scholar] [CrossRef] [PubMed]
- Patra, A.K. Urea/Ammonia metabolism in the rumen and toxicity in ruminants. In Rumen Microbiology: From Evolution to Revolution; Puniya, A.K., Singh, R., Kamra, D.N., Eds.; Springer: New Delhi, India, 2015; pp. 329–341. [Google Scholar]
- Shriver, B.J.; Hoover, W.H.; Sargent, J.P.; Crawford, R.J., Jr.; Thayne, W.V. Fermentation of a high concentrate diet as affected by ruminal ph and digesta flow. J. Dairy Sci. 1986, 69, 413–419. [Google Scholar] [CrossRef]
- Zhao, C.; Wang, Y.; Peng, Z.; Sun, X.; Sun, G.; Yuan, X.; Li, X.; Liu, G. Subacute ruminal acidosis suppressed the expression of MCT1 in rumen of cows. J. Cell. Physiol. 2019, 234, 11734–11745. [Google Scholar] [CrossRef]
- Sheng, L.; Jena, P.K.; Hu, Y.; Liu, H.; Nagar, N.; Kalanetra, K.M.; French, S.W.; French, S.W.; Mills, D.A.; Wan, Y.Y. Hepatic inflammation caused by dysregulated bile acid synthesis is reversible by butyrate supplementation. J. Pathol. 2017, 243, 431–441. [Google Scholar] [CrossRef] [Green Version]
- Peng, C.; Lin, H.; Chung, D.; Huang, C.; Wang, C. Mulberry leaf extracts prevent obesity-induced NAFLD with regulating adipocytokines, inflammation and oxidative stress. J. Food Drug Anal. 2018, 26, 778–787. [Google Scholar] [CrossRef]
- Urai, T.; Haryanto; Mukai, K.; Matsushita, T.; Asano, K.; Nakajima, Y.; Okuwa, M.; Sugama, J.; Nakatani, T. The relationship between cutaneous wounds made on obese mice or those with decreased body weight and serum leptin level. Health 2016, 8, 1015–1028. [Google Scholar] [CrossRef] [Green Version]
- Hwang, J.; Shieh, D.; Shyu, Y.; Hsu, C.; Lin, C. Antidiabetic, antihyperlipidemic, and antioxidant activities of mulberry lees fermented products in diabetic mice. J. Food Sci. 2018, 83, 2866–2872. [Google Scholar] [CrossRef]
- Henderson, G.; Cox, F.; Ganesh, S.; Jonker, A.; Young, W.; Janssen, P.H. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 2015, 5, 14567. [Google Scholar] [CrossRef] [PubMed]
- Shade, A.; Handelsman, J. Beyond the Venn diagram: The hunt for a core microbiome. Environ. Microbiol. 2012, 14, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Liu, J.; Feng, P.; Zhu, W.; Mao, S. Grain-rich diets altered the colonic fermentation and mucosa-associated bacterial communities and induced mucosal injuries in goats. Sci. Rep. 2016, 6, 20329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derakhshani, H.; Tun, H.M.; Cardoso, F.C.; Plaizier, J.C.; Khafipour, E.; Loor, J.J. Linking peripartal dynamics of ruminal microbiota to dietary changes and production parameters. Front. Microbiol. 2017, 7, 2143. [Google Scholar] [CrossRef] [Green Version]
- Golder, H.M.; Denman, S.E.; Mcsweeney, C.; Wales, W.J.; Auldist, M.J.; Wright, M.M.; Marett, L.C.; Greenwood, J.S.; Hannah, M.C.; Celi, P. Effects of partial mixed rations and supplement amounts on milk production and composition, ruminal fermentation, bacterial communities, and ruminal acidosis. J. Dairy Sci. 2014, 97, 5763–5785. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Dong, D.; Wang, H.; Müller, K.; Qin, Y.; Wang, H.; Wu, W. Metagenomic analysis of microbial consortia enriched from compost: New insights into the role of Actinobacteria in lignocellulose decomposition. Biotechnol. Biofuels 2016, 9, 22. [Google Scholar] [CrossRef] [Green Version]
- Tan, N.D.; Wanapat, M.; Uriyapongson, S.; Cherdthong, A.; Pilajun, R. Enhancing mulberry leaf meal with urea by pelleting to improve rumen fermentation in cattle. Asian Austral. J. Anim. 2012, 25, 452–461. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Mao, X.; He, J.; Yu, B.; Huang, Z.; Yu, J.; Zheng, P.; Chen, D. Dietary fibre affects intestinal mucosal barrier function and regulates intestinal bacteria in weaning piglets. Brit. J. Nutr. 2013, 110, 1837–1848. [Google Scholar] [CrossRef] [Green Version]
- Uyeno, Y.; Shigemori, S.; Shimosato, T. Effect of probiotics/prebiotics on cattle health and productivity. Microbes Environ. 2015, 30, 126–132. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Liu, F.; Liu, J.; Liao, S.; Zou, Y. Mulberry leaf polyphenols and fiber induce synergistic antiobesity and display a modulation effect on gut microbiota and metabolites. Nutrients 2019, 11, 1017. [Google Scholar] [CrossRef] [Green Version]
- Chávez-Carbajal, A.; Nirmalkar, K.; Pérez-Lizaur, A.; Hernández-Quiroz, F.; Ramírez-del-Alto, S.; García-Mena, J.; Hernández-Guerrero, C. Gut microbiota and predicted metabolic pathways in a sample of Mexican women affected by obesity and obesity plus metabolic syndrome. Int. J. Mol. Sci. 2019, 20, 438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez, N.; Abdalla, A.L.; Galindo, J.; Santos, M.R.; Louvandini, P.; Louvandini, H. Effect of five inclusion levels of mulberry (Morus alba cv. cubana) on methanogens and some main cellulolytic populations within rumen liquor of water buffalos (Bubalus bubalis). Cuba. J. Agric. Sci. 2016, 50, 393–402. [Google Scholar]
- Belanche, A.; Jones, E.; Parveen, I.; Newbold, C.J. A Metagenomics approach to evaluate the impact of dietary supplementation with Ascophyllum nodosum or Laminaria digitate on rumen function in rusitec fermenters. Front. Microbiol. 2016, 7, 299. [Google Scholar] [CrossRef]
- Ramayo-Caldas, Y.; Mach, N.; Lepage, P.; Levenez, F.; Denis, C.; Lemonnier, G.; Leplat, J.; Billon, Y.; Berri, M.; Doré, J.; et al. Phylogenetic network analysis applied to pig gut microbiota identifies an ecosystem structure linked with growth traits. ISME J. 2016, 10, 2973–2977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Y.; Malmuthuge, N.; Steele, M.A.; Guan, L.L. Shift of hindgut microbiota and microbial short chain fatty acids profiles in dairy calves from birth to pre-weaning. FEMS Microbiol. Ecol. 2018, 94. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liao, X.; Sparks, J.B.; Luo, X.M. Dynamics of gut microbiota in autoimmune lupus. Appl. Environ. Microb. 2014, 80, 7551–7560. [Google Scholar] [CrossRef] [Green Version]
- Jordan, F.; Lauria, M.; Scotti, M.; Nguyen, T.P.; Praveen, P.; Morine, M.; Priami, C. Diversity of key players in the microbial ecosystems of the human body. Sci. Rep. 2015, 5, 15920. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, H.; Wu, X.; Shang, S.; Yan, J.; Chen, Y.; Zhang, H.; Tang, X. Characterization of the gut microbiota in the golden takin (Budorcas taxicolor bedfordi). AMB Express. 2017, 7, 81. [Google Scholar] [CrossRef] [Green Version]
- Clemmons, B.A.; Martino, C.; Powers, J.B.; Campagna, S.R.; Voy, B.H.; Donohoe, D.; Gaffney, J.; Embree, M.; Myer, P.R. Rumen bacteria and serum metabolites predictive of feed efficiency phenotypes in beef cattle. Sci. Rep. 2019, 9, 19265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plöger, S.; Stumpff, F.; Penner, G.B.; Schulzke, J.D.; Gäbel, G.; Martens, H.; Shen, Z.; Günzel, D.; Aschenbach, J.R. Microbial butyrate and its role for barrier function in the gastrointestinal tract. Ann. N. Y. Acad. Sci. 2012, 1258, 52–59. [Google Scholar] [CrossRef] [Green Version]
- Peng, Z.; Zhang, J.; Fanning, S.; Wang, L.; Li, M.; Maheshwari, N.; Sun, J.; Li, F. Effects of metal and metalloid pollutants on the microbiota composition of feces obtained from twelve commercial pig farms across China. Sci. Total Environ. 2019, 647, 577–586. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, L.; Liu, J.; Zhu, W.; Mao, S. A high grain diet dynamically shifted the composition of mucosa-associated microbiota and induced mucosal injuries in the colon of sheep. Front. Microbiol. 2017, 8, 2080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, J.; Yi, L.; Hai, L.; Ming, L.; Gao, W.; Ji, R. Characterizing the bacterial microbiota in different gastrointestinal tract segments of the Bactrian camel. Sci. Rep. 2018, 8, 654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamendella, R.; Domingo, J.W.; Ghosh, S.; Martinson, J.; Oerther, D.B. Comparative fecal metagenomics unveils unique functional capacity of the swine gut. BMC Microbiol. 2011, 11, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Mao, H.; Wang, Z.; Tian, Y. Succession of organics metabolic function of bacterial community in swine manure composting. J. Hazard. Mater. 2018, 360, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Makela, M.R.; Difalco, M.; Mcdonnell, E.; Nguyen, T.T.M.; Wiebenga, A.; Hilden, K.; Peng, M.; Grigoriev, I.V.; Tsang, A.; De Vries, R.P. Genomic and exoproteomic diversity in plant biomass degradation approaches among Aspergilli. Stud. Mycol. 2018, 91, 79–99. [Google Scholar] [CrossRef]
- Afrin, T.; Murase, K.; Kounosu, A.; Hunt, V.L.; Bligh, M.; Maeda, Y.; Hino, A.; Maruyama, H.; Tsai, I.J.; Kikuchi, T. Sequential changes in the host gut microbiota during infection with the intestinal parasitic nematode Strongyloides venezuelensis. Front. Cell. Infect. Mi. 2019, 9, 217. [Google Scholar] [CrossRef] [Green Version]
- Kaakoush, N.O.; Martire, S.I.; Raipuria, M.; Mitchell, H.M.; Nielsen, S.; Westbrook, R.F.; Morris, M.J. Alternating or continuous exposure to cafeteria diet leads to similar shifts in gut microbiota compared to chow diet. Mol. Nutr. Food Res. 2017, 61, 1500815. [Google Scholar] [CrossRef] [Green Version]
- Meale, S.J.; Li, S.C.; Azevedo, P.; Derakhshani, H.; DeVries, T.J.; Plaizier, J.C.; Steele, M.A.; Khafipour, E. Weaning age influences the severity of gastrointestinal microbiome shifts in dairy calves. Sci. Rep. 2017, 7, 198. [Google Scholar] [CrossRef]







| Groups | R | p-Value | |||||
|---|---|---|---|---|---|---|---|
| Supplemental levels | S (S1,S2) | Phylum | Genus | OTU | Phylum | Genus | OTU |
| −0.074 | −0.259 | −0.185 | 0.714 | 1.000 | 1.000 | ||
| E (E1,E2,E3) | Phylum | Genus | OTU | Phylum | Genus | OTU | |
| 0.111 | 0.243 | 0.350 | 0.265 | 0.122 | 0.055 | ||
| Feed types | T a(C, S, E) | Phylum | Genus | OTU | Phylum | Genus | OTU |
| 0.328 | 0.292 | 0.336 | 0.009 | 0.015 | 0.01 | ||
| Taxa a | Treatment | SEM | p | ||
|---|---|---|---|---|---|
| C | S | E | |||
| Anaerobic fungi | 6.30 | 4.20 | 5.43 | 0.330 | 0.058 |
| Methanogens | 4.30 | 5.31 | 5.52 | 0.187 | 0.066 |
| Ciliate protozoa | 7.17 | 7.17 | 6.83 | 0.151 | 0.568 |
| Item | Treatment | SEM | p | ||||
|---|---|---|---|---|---|---|---|
| C | S | E | |||||
| Rumen morphology | PL a(μm) | 747.81 | 973.86 | 729.26 | 61.312 | 0.186 | |
| PW b(μm) | 301.89 | 266.44 | 245.01 | 10.088 | 0.127 | ||
| PSA c(μm2) | 0.13 | 0.22 | 0.11 | 0.197 | 0.423 | ||
| RWT d(μm) | 1545.73 | 1444.46 | 1426.49 | 74.591 | 0.865 | ||
| Rumen fermentation parameters | pH | 6.37 | 6.00 | 6.39 | 0.086 | 0.097 | |
| VFA | Acetate(A) (%) | 64.27 | 57.13 | 60.69 | 1.046 | 0.053 | |
| Propionate (P) (%) | 18.17 | 21.13 | 19.01 | 0.853 | 0.435 | ||
| Butyrate (%) | 12.36 | 14.90 | 14.85 | 0.827 | 0.557 | ||
| Isobutyrate (%) | 1.32 | 1.10 | 1.46 | 0.109 | 0.345 | ||
| Valerate (%) | 1.42 | 2.38 | 1.43 | 0.296 | 0.336 | ||
| Isovalerate (%) | 2.46 | 2.26 | 2.55 | 0.254 | 0.884 | ||
| A:P | 3.54 | 2.70 | 3.19 | 0.164 | 0.375 | ||
| Total VFA (mmol/L) | 112.67 | 123.60 | 95.59 | 7.004 | 0.199 | ||
| Ammonium nitrogen(mg/dL) | 29.38 | 33.65 | 27.23 | 2.468 | 0.536 | ||
| CMCase activity e(U/mL) | 3.86 | 3.46 | 4.10 | 0.472 | 0.847 | ||
| Items a | Treatment | SEM | p | ||
|---|---|---|---|---|---|
| C | S | E | |||
| TC (mmol/L) | 2.27 | 2.34 | 2.57 | 0.098 | 0.439 |
| TG (mmol/L) | 0.25 | 0.37 | 0.25 | 0.026 | 0.109 |
| HDL-C (mmol/L) | 1.45 | 1.31 | 1.39 | 0.047 | 0.583 |
| LDL-C (mmol/L) | 0.62 | 0.51 | 0.60 | 0.030 | 0.356 |
| LPS (U/L) | 53.17 | 68.50 | 59.05 | 3.710 | 0.347 |
| LPL (μmol/mL h) | 2.19 | 2.11 | 2.07 | 0.119 | 0.948 |
| Leptin (ng/mL) | 7.26 | 4.75 | 6.81 | 0.342 | 0.003 |
| FASN (ng/mL) | 7.97 | 7.30 | 7.74 | 0.331 | 0.780 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Shen, Q.; Zhong, S.; Chen, Y.; Yang, Y. Comparison of Rumen Microbiota and Serum Biochemical Indices in White Cashmere Goats Fed Ensiled or Sun-Dried Mulberry Leaves. Microorganisms 2020, 8, 981. https://doi.org/10.3390/microorganisms8070981
Wang Y, Shen Q, Zhong S, Chen Y, Yang Y. Comparison of Rumen Microbiota and Serum Biochemical Indices in White Cashmere Goats Fed Ensiled or Sun-Dried Mulberry Leaves. Microorganisms. 2020; 8(7):981. https://doi.org/10.3390/microorganisms8070981
Chicago/Turabian StyleWang, Yaoyue, Qingmiao Shen, Shu Zhong, Yulin Chen, and Yuxin Yang. 2020. "Comparison of Rumen Microbiota and Serum Biochemical Indices in White Cashmere Goats Fed Ensiled or Sun-Dried Mulberry Leaves" Microorganisms 8, no. 7: 981. https://doi.org/10.3390/microorganisms8070981