Microfluidic-Based Approaches for Foodborne Pathogen Detection
Abstract
:1. Introduction
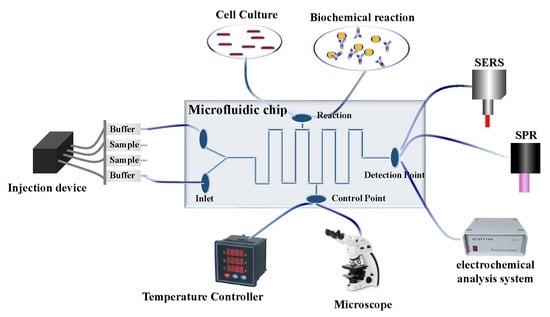
2. Microfluidic Chips
3. Sample Preparation in Microfluidics
3.1. For Single Component
3.2. Complex Components in Food Matrix
3.2.1. Special Materials and Sampling Methods
3.2.2. Bio-Recognition Molecules
4. Application of Microfluidic Combined with Different Technologies
4.1. Biosensor-Based Microfluidics for the Detection of Foodborne Pathogens
4.1.1. Microfluidic Chips with Optical Detection
Surface Plasmon Resonance (SPR) Biosensors
Optical Fibre Biosensors
4.1.2. Microfluidic Chip with Electrochemical Detection
4.2. Immunoassay-Based Microfluidics for the Detection of Foodborne Pathogens
4.2.1. Enzyme-Linked Immunosorbent Assay (ELISA)
4.2.2. Immunomagnetic Fluorescence Assay (IMS)
4.3. Nucleic Acid-Based Microfluidics for the Detection of Foodborne Pathogens
4.3.1. Polymerase Chain Reaction (PCR)
4.3.2. Multiplex PCR
4.3.3. Loop-Mediated Isothermal Amplification (LAMP)
5. Challenges and Opportunities
6. Conclusions
Funding
Conflicts of Interest
References
- Chapman, B.; Gunter, C. Local Food Systems Food Safety Concerns. Microbiol. Spectr. 2018, 6, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Li, M.; Xu, Z. Detection of Foodborne Pathogens by Surface Enhanced Raman Spectroscopy. Front. Microbiol. 2018, 9, 1236. [Google Scholar] [CrossRef] [PubMed]
- Jones, T.F.; Yackley, J. Foodborne Disease Outbreaks in the United States: A Historical Overview. Foodborne Pathog. Dis. 2018, 15, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Parisi, A.; Crump, J.A.; Glass, K.; Howden, B.P.; Furuya-Kanamori, L.; Vilkins, S.; Gray, D.J.; Kirk, M.D. Health Outcomes from Multidrug-Resistant Salmonella Infections in High-Income Countries: A Systematic Review and Meta-Analysis. Foodborne Pathog. Dis. 2018, 15, 428–436. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Lin, C.W.; Wang, J.; Oh, D.H. Advances in rapid detection methods for foodborne pathogens. J. Microbiol. Biotechnol. 2014, 24, 297. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Y.; Zhou, R.; Li, L.; Peters, B.M.; Li, B.; Lin, C.W.; Peters, B.M.; Chuang, T.L.; Chen, D.Q.; Zhao, X.H.; et al. Viable but non-culturable state and toxin gene expression of enterohemorrhagic Escherichia coli O157 under cryopreservation. Res. Microbiol. 2017, 168, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Torgerson, P.R.; Devleesschauwer, B.; Praet, N.; Speybroeck, N.; Willingham, A.L.; Kasuga, F.; Rokni, M.B.; Zhou, X.N.; Fèvre, E.M.; Sripa, B.; et al. World Health Organization Estimates of the Global and Regional Disease Burden of 11 Foodborne Parasitic Diseases, 2010: A Data Synthesis. PLoS Med. 2015, 12, e1001920. [Google Scholar] [CrossRef]
- Yang, S.C.; Lin, C.H.; Aljuffali, I.A.; Fang, J.Y. Current pathogenic Escherichia coli foodborne outbreak cases and therapy development. Arch. Microbiol. 2017, 199, 811–825. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.H.; Wei, C.J.; Zhong, J.L.; Jin, S.W. Research advance in rapid detection of foodborne Staphylococcus aureus. Biotechnol. Biotechnol. Equip. 2016, 30, 827–833. [Google Scholar] [CrossRef]
- Umesha, S.; Manukumar, H.M. Advanced Molecular Diagnostic Techniques for Detection of Food-borne Pathogens: Current Applications and Future Challenges. Crit. Rev. Food Sci. Nutr. 2018, 58, 84–104. [Google Scholar] [CrossRef]
- Wei, C.J.; Zhong, J.L.; Hu, T.; Zhao, X.H. Simultaneous detection of Escherichia coli O157:H7, Staphylococcus aureus and Salmonella by multiplex PCR in milk. 3 Biotech 2018, 8, 76. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhao, F.; Wang, J.; Zhong, N. Biofilm formation and control strategies of foodborne pathogens: Food safety perspectives. RSC Adv. 2017, 7, 36670–36683. [Google Scholar] [CrossRef]
- Zhong, J.; Zhao, X. Isothermal amplification technologies for the detection of foodborne pathogens. Food Anal. Methods 2018, 11, 1543–1560. [Google Scholar] [CrossRef]
- Zhao, X.; Zhong, J.; Wei, C.; Lin, C.W.; Ding, T. Current perspectives on viable but non-culturable state in foodborne pathogens. Front. Microbiol. 2017, 8, 580. [Google Scholar] [CrossRef] [PubMed]
- Holmes, D.; Gawad, S. The application of microfluidics in biology. Methods Mol. Biol. 2010, 583, 55–80. [Google Scholar] [PubMed]
- Yujie, L.I.; Huo, Y.; Di, L.I.; Tang, X.; Shi, F.; Wang, C. Technology, application and development of microfluidics. J. Hebei Univ. Sci. Technol. 2014, 35, 11. [Google Scholar]
- Wen, N.; Zhao, Z.; Fan, B.; Chen, D.; Men, D.; Wang, J.; Chen, J. Development of Droplet Microfluidics Enabling High-Throughput Single-Cell Analysis. Molecules 2016, 21, 881. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.-K.; Ke, Y.; Jun, Z.; Can-Can, Z.; Ling, Z.; Yong, L. Rapid Detection of Hepatitis B Virus Nucleic Acid Based on Microfluidic Chip Using Fluorescence Quantitative PCR. J. Anal. Sci. 2018, 34, 11–15. [Google Scholar]
- Wang, C.; Madiyar, F.; Yu, C.; Li, J. Detection of extremely low concentration waterborne pathogen using a multiplexing self-referencing SERS microfluidic biosensor. J. Biol. Eng. 2017, 11, 9. [Google Scholar] [CrossRef]
- Wan, L.; Chen, T.; Gao, J.; Dong, C.; Wong, A.H.; Jia, Y.; Mak, P.; Deng, C.X.; Martins, R.P. A digital microfluidic system for loop-mediated isothermal amplification and sequence specific pathogen detection. Sci. Rep. 2017, 7, 14586. [Google Scholar] [CrossRef]
- Jokerst, J.C.; Adkins, J.A.; Bisha, B.; Mentele, M.M.; Goodridge, L.D.; Henry, C.S. Development of a Paper-Based Analytical Device for Colorimetric Detection of Select Foodborne Pathogens. Anal. Chem. 2012, 84, 2900–2907. [Google Scholar] [CrossRef] [PubMed]
- Long, H.; Bao, L.J.; Habeeb, A.A.; Lu, P.X. Effects of doping concentration on the surface plasmonic resonances and optical nonlinearities in AGZO nano-triangle arrays. Opt. Quantum Electron. 2017, 49, 345. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, D.M.; Lu, P.; Sun, Q.Z.; Yang, W.; Wang, S.; Liu, L.; Zhang, J.S. Dual-Parameters Optical Fiber Sensor with Enhanced Resolution Using Twisted MMF Based on SMS Structure. IEEE Sens. J. 2017, 17, 3045–3051. [Google Scholar] [CrossRef]
- Squires, T.M.; Quake, S.R. Microfluidics: Fluid physics at the nanoliter scale. Rev. Mod. Phys. 2005, 77, 977–1026. [Google Scholar] [CrossRef] [Green Version]
- Whitesides, G.M. The origins and the future of microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Weng, X.; Neethirajan, S. Paper-based microfluidic aptasensor for food safety. J. Food Saf. 2017, 38, e12412. [Google Scholar] [CrossRef]
- Xu, J.; Kawano, H.; Liu, W.W.; Hanada, Y.; Lu, P.X.; Miyawaki, A.; Midorikawa, K.; Sugioka, K. Controllable alignment of elongated microorganisms in 3D microspace using electrofluidic devices manufactured by hybrid femtosecond laser microfabrication. Microsyst. Nanoeng. 2017, 3, 16078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Z.Q.; He, X.Q.; Liew, K.M. A sensitive interval of imperfect interface parameters based on the analysis of general solution for anisotropic matrix containing an elliptic inhomogeneity. Int. J. Solids Struct. 2015, 73–74, 67–77. [Google Scholar] [CrossRef]
- Hou, M.; Wang, Y.; Liu, S.; Guo, J.; Li, Z.; Lu, P. Sensitivity-Enhanced Pressure Sensor with Hollow-Core Photonic Crystal Fiber. J. Lightwave Technol. 2014, 32, 4637–4641. [Google Scholar]
- Manz, A.; Graber, N.; Widmer, H.M. Miniaturized total chemical analysis systems: A novel concept for chemical sensing. Sens. Actuators B Chem. 1990, 1, 244–248. [Google Scholar] [CrossRef]
- Manz, A.; Harrison, D.J.; Verpoorte, E.M.J.; Fettinger, J.C.; Paulus, A.; Lüdi, H.; Widmer, H.M. Planar chips technology for miniaturization and integration of separation techniques into monitoring systems: Capillary electrophoresis on a chip. J. Chromatogr. A 1992, 593, 253–258. [Google Scholar] [CrossRef]
- Woolley, A.T.; Mathies, R.A. Ultra-high-speed DNA sequencing using capillary electrophoresis chips. Anal. Chem. 1995, 67, 3676–3680. [Google Scholar] [CrossRef] [PubMed]
- Woolley, A.T.; Hadley, D.; Landre, P.; Demello, A.J.; Mathies, R.A.; Northrup, M.A. Functional integration of PCR amplification and capillary electrophoresis in a microfabricated DNA analysis device. Anal. Chem. 1996, 68, 4081–4086. [Google Scholar] [CrossRef] [PubMed]
- Brahmasandra, S.N.; Johnson, B.N.; Webster, J.R.; Burke, D.T.; Mastrangelo, C.H.; Burns, M.A. On-chip DNA band detection in microfabricated separation systems. Proc. Spie—Int. Soc. Opt. Eng. 1998, 3515, 242–251. [Google Scholar]
- Anderson, J.R.; Chiu, D.T.; Jackman, R.J.; Cherniavskaya, O.; Mcdonald, J.C.; Wu, H.; Whitesides, S.H.; Whitesides, G.M. Fabrication of topologically complex three-dimensional microfluidic systems in PDMS by rapid prototyping. Anal. Chem. 2000, 72, 3158–3164. [Google Scholar] [CrossRef] [PubMed]
- Ghaemmaghami, A.M.; Hancock, M.J.; Harrington, H.; Kaji, H.; Khademhosseini, A. Biomimetic tissues on a chip for drug discovery. Drug Discov. Today 2012, 17, 173–181. [Google Scholar] [CrossRef] [Green Version]
- Marre, S.; Jensen, K.F. Synthesis of micro and nanostructures in microfluidic systems. Chem. Soc. Rev. 2010, 39, 1183–1202. [Google Scholar] [CrossRef] [PubMed]
- Mu, X.; Zheng, W.; Sun, J.; Zhang, W.; Jiang, X. Microfluidics for Manipulating Cells. Small 2013, 9, 9–21. [Google Scholar] [CrossRef]
- Zhao, D.; Liu, W.W.; Ke, S.L.; Liu, Q.J. Large lateral shift in complex dielectric multilayers with nearly parity-time symmetry. Opt. Quantum Electron. 2018, 50, 323. [Google Scholar] [CrossRef]
- Chuang, T.L.; Chang, C.C.; Chu-Su, Y.; Wei, S.C.; Zhao, X.H.; Hsueh, P.R.; Lin, C.W. Disposable surface plasmon resonance aptasensor with membranebased sample handling design for quantitative interferon-gamma detection. Lab Chip 2014, 14, 2968–2977. [Google Scholar] [CrossRef]
- Atsushi, K.; Akiko, I.; Tamotsu, Y.; Yoshiaki, U.; Eiichi, T.; Yuzuru, T. Highly sensitive elemental analysis for Cd and Pb by liquid electrode plasma atomic emission spectrometry with quartz glass chip and sample flow. Anal. Chem. 2011, 83, 9424–9430. [Google Scholar]
- Francisca, A.; Neha, S.; Mohammad-Ali, S.; Dongfei, L.; Bárbara, H.B.; Makila, E.M.; Jarno, J.S.; Jouni, T.H.; Pedro, L.G.; Bruno, S.; et al. Microfluidic Assembly of a Multifunctional Tailorable Composite System Designed for Site Specific Combined Oral Delivery of Peptide Drugs. Acs Nano 2015, 9, 8291–8302. [Google Scholar]
- Xuan, T.V.; Stockmann, R.; Wolfrum, B.; Offenhäusser, A.; Ingebrandt, S. Fabrication and application of a microfluidic-embedded silicon nanowire biosensor chip. Phys. Status Solidi 2010, 207, 850–857. [Google Scholar]
- Zhang, H.; Dongfei, L.; Mohammad-Ali, S.; Ermei, M.K.; Bárbara, H.B.; Jarno, S.; Hirvonen, J.; Santos, H.A. Fabrication of a multifunctional nano-in-micro drug delivery platform by microfluidic templated encapsulation of porous silicon in polymer matrix. Adv. Mater. 2014, 26, 4497–4503. [Google Scholar] [CrossRef] [PubMed]
- Crabtree, H.J.; Morrissey, Y.C.; Taylor, B.J.; Liang, T.; Johnstone, R.W.; Stickel, A.J.; Manage, P.; Atrazhev, A.; Backhouse, C.J.; Pilarski, L.M. Inhibition of on-chip PCR using PDMS–glass hybrid microfluidic chips. Microfluid. Nanofluid. 2012, 13, 383–398. [Google Scholar] [CrossRef]
- Tan, F.; Leung, P.H.M.; Liu, Z.B.; Zhang, Y.; Xiao, L.; Ye, W.; Zhang, X.; Yi, L.; Yang, M. A PDMS microfluidic impedance immunosensor for E. coli O157:H7 and Staphylococcus aureus detection via antibody-immobilized nanoporous membrane. Sens. Actuators B Chem. 2011, 159, 328–335. [Google Scholar] [CrossRef]
- Al-Shehri, S.; Palitsin, V.; Webb, R.P.; Grime, G.W. Fabrication of three-dimensional SU-8 microchannels by proton beam writing for microfluidics applications: Fluid flow characterisation. Nucl. Instrum. Methods Phys. Res. B 2015, 348, 223–228. [Google Scholar] [CrossRef] [Green Version]
- Dy, A.J.; Cosmanescu, A.; Sluka, J.; Glazier, J.A.; Stupack, D.; Amarie, D. Fabricating microfluidic valve master molds in SU-8 photoresist. J. Micromech. Microeng. 2014, 24, 057001. [Google Scholar] [CrossRef] [Green Version]
- Floquet, C.F.A.; Sieben, V.J.; Milani, A.; Joly, E.P.; Ogilvie, I.R.G.; Morgan, H.; Mowlem, M.C. Nanomolar detection with high sensitivity microfluidic absorption cells manufactured in tinted PMMA for chemical analysis. Talanta 2011, 84, 235–239. [Google Scholar] [CrossRef]
- Wu, N.; Zhu, Y.G.; Brown, S.; Oakeshott, J.; Peat, T.; Surjadi, R.; Easton, C.; Leech, P.W.; Sexton, B.A. A PMMA microfluidic droplet platform for in Vitro protein expression using crude E. Coli S30 extract. Lab Chip 2009, 9, 3391–3398. [Google Scholar] [CrossRef]
- Stojkovič, G.; Krivec, M.; Vesel, A.; Marinšek, M.; Žnidaršič-Plazl, P. Surface cell immobilization within perfluoroalkoxy microchannels. Appl. Surf. Sci. 2014, 320, 810–817. [Google Scholar] [CrossRef]
- Detlev, B.; Alfred, D.; Frank, K.; Martin, L. Poly (vinyl alcohol)-coated microfluidic devices for high-performance microchip electrophoresis. Electrophoresis 2015, 23, 3567–3573. [Google Scholar]
- Huang, K.W.; Wu, Y.C.; Lee, J.A.; Chiou, P.Y. Microfluidic integrated optoelectronic tweezers for single-cell preparation and analysis. Lab Chip 2013, 13, 3721–3727. [Google Scholar] [CrossRef] [PubMed]
- Hong, B.; Xue, P.; Wu, Y.; Bao, J.; Chuah, Y.J.; Kang, Y. A concentration gradient generator on a paper-based microfluidic chip coupled with cell culture microarray for high-throughput drug screening. Biomed. Microdevices 2016, 18, 21. [Google Scholar] [CrossRef] [PubMed]
- Li, W.L.; Wu, A.; Li, Z.C.; Zhang, G.; Yu, W.Y. A new calibration method between an optical sensor and a rotating platform in turbine blade inspection. Meas. Sci. Technol. 2017, 28, 035009. [Google Scholar] [CrossRef]
- Zeng, D.; Chen, Z.; Jiang, Y.; Xue, F.; Li, B. Advances and Challenges in Viability Detection of Foodborne Pathogens. Front. Microbiol. 2016, 7, 1833. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Hai-Qing, G.; Xudong, Z.; Chaoping, L.; Chunchau, S. A microfluidic liquid phase nucleic acid purification chip to selectively isolate DNA or RNA from low copy/single bacterial cells in minute sample volume followed by direct on-chip quantitative PCR assay. Anal. Chem. 2013, 85, 1484–1491. [Google Scholar] [CrossRef]
- Wu, J.; Kodzius, R.; Cao, W.; Wen, W. Extraction, amplification and detection of DNA in microfluidic chip-based assays. Microchim. Acta 2014, 181, 1611–1631. [Google Scholar] [CrossRef]
- Tachibana, H.; Saito, M.; Shibuya, S.; Tsuji, K.; Miyagawa, N.; Yamanaka, K.; Tamiya, E. On-chip quantitative detection of pathogen genes by autonomous microfluidic PCR platform. Biosens. Bioelectron. 2015, 74, 725–730. [Google Scholar] [CrossRef]
- Wang, Y.; Jianfeng, P.; Zunzhong, Y.; Jian, W.; Yibin, Y. Impedimetric immunosensor based on gold nanoparticles modified graphene paper for label-free detection of Escherichia coli O157:H7. Biosens. Bioelectron. 2013, 4, 492–498. [Google Scholar] [CrossRef]
- Cate, D.M.; Adkins, J.A.; Mettakoonpitak, J.; Henry, C.S.; Chem, A. Recent Developments in Paper-Based Microfluidic Devices. Anal. Chem. 2015, 87, 19–41. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.C.; Wang, Y.N.; Fu, L.M.; Chen, K.L. Microfluidic paper-based chip platform for benzoic acid detection in food. Food Chem. 2018, 249, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.F.; Juan, H.U.; Zheng, G.; Zhao, G.H. Application of microfluidic chip in food safety analysis. Sci. Technol. Food Ind. 2011, 32, 401–404. [Google Scholar]
- Zheng, Y.; Mao, S.; Liu, S.; Wong, S.H.; Wang, Y.W. Normalized Relative RBC-Based Minimum Risk Bayesian Decision Approach for Fault Diagnosis of Industrial Process. IEEE Trans. Ind. Electron. 2016, 63, 7723–7732. [Google Scholar] [CrossRef]
- Ganesh, I.; Tran, B.M.; Kim, Y.; Kim, J.; Cheng, H.; Lee, N.Y.; Park, S. An integrated microfluidic PCR system with immunomagnetic nanoparticles for the detection of bacterial pathogens. Biomed. Microdevices 2016, 18, 116. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.J.; Park, B.H.; Choi, G.; Seo, J.H.; Jung, J.H.; Choi, J.S.; Kim, D.H.; Seo, T.S. Fully automated and colorimetric foodborne pathogen detection on an integrated centrifugal microfluidic device. Lab Chip 2016, 16, 1917–1926. [Google Scholar] [CrossRef]
- Sayad, A.; Ibrahim, F.; Mukim, S.U.; Cho, J.; Madou, M.; Thong, K.L. A microdevice for rapid, monoplex and colorimetric detection of foodborne pathogens using a centrifugal microfluidic platform. Biosens. Bioelectron. 2017, 100, 96–104. [Google Scholar] [CrossRef]
- Li, X.; Ximenes, E.; Amalaradjou, M.A.; Vibbert, H.B.; Foster, K.; Jones, J.; Liu, X.Y.; Bhunia, A.K.; Ladisch, M.R. Rapid sample processing for detection of food-borne pathogens via cross-flow microfiltration. Appl. Environ. Microbiol. 2013, 79, 7048–7054. [Google Scholar] [CrossRef]
- Shu, B.; Zhang, C.; Xing, D. Segmented continuous-flow multiplex polymerase chain reaction microfluidics for high-throughput and rapid foodborne pathogen detection. Anal. Chim. Acta 2014, 826, 51–60. [Google Scholar] [CrossRef]
- Savas, S.; Ersoy, A.; Gulmez, Y.; Kilic, S.; Levent, B.; Altintas, Z. Nanoparticle Enhanced Antibody and DNA Biosensors for Sensitive Detection of Salmonella. Materials 2018, 11, 1541. [Google Scholar] [CrossRef]
- Wu, S.; Duan, N.; Shi, Z.; Fang, C.; Wang, Z. Simultaneous aptasensor for multiplex pathogenic bacteria detection based on multicolor upconversion nanoparticles labels. Anal. Chem. 2014, 86, 3100–3107. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.H.; Um, E.; Diaz, A.; Driscoll, H.; Rodas, M.J.; Domansky, K.; Rodas, M.J.; Watters, A.L.; Super, M.; Stone, H.A.; et al. Optimization of Pathogen Capture in Flowing Fluids with Magnetic Nanoparticles. Small 2015, 11, 5657–5666. [Google Scholar] [CrossRef] [PubMed]
- Dao, T.N.T.; Yoon, J.; Jin, C.E.; Koo, B.; Han, K.; Yong, S.; Lee, T.Y. Rapid and Sensitive Detection of Salmonella based on Microfluidic Enrichment with a Label-free Nanobiosensing Platform. Sens. Actuators B Chem. 2017, 262, 588–594. [Google Scholar] [CrossRef]
- Deshmukh, R.A.; Joshi, K.; Bhand, S.; Roy, U. Recent developments in detection and enumeration of waterborne bacteria: A retrospective minireview. Microbiologyopen 2016, 5, 901–922. [Google Scholar] [CrossRef] [PubMed]
- Law, J.W.; Ab Mutalib, N.S.; Chan, K.G.; Lee, L.H. Rapid methods for the detection of foodborne bacterial pathogens: Principles, applications, advantages and limitations. Front. Microbiol. 2015, 5, 770. [Google Scholar] [CrossRef] [PubMed]
- Ríos, Á.; Zougagh, M. Modern qualitative analysis by miniaturized and microfluidic systems. Trends Anal. Chem. 2015, 69, 105–113. [Google Scholar] [CrossRef]
- Kant, K.; Shahbazi, M.A.; Dave, V.P.; Ngo, T.A.; Chidambara, V.A.; Linh, Q.T.; Dang, D.B.; Anders, W. Microfluidic devices for sample preparation and rapid detection of foodborne pathogens. Biotechnol. Adv. 2018, 36, 1003–1024. [Google Scholar] [CrossRef] [Green Version]
- Chi, L.W.; Olivo, M. Surface Plasmon Resonance Imaging Sensors: A Review. Plasmonics 2014, 9, 809–824. [Google Scholar]
- Lee, H.; Xu, L.; Koh, D.; Nyayapathi, N.; Oh, K.W. Various on-chip sensors with microfluidics for biological applications. Sensors 2014, 14, 17008–17036. [Google Scholar] [CrossRef]
- Liu, S.H.; Tian, J.; Liu, N.L.; Lu, P.X. Temperature Insensitive Liquid Level Sensor Based on Antiresonant Reflecting Guidance in Silica Tube. J. Lightwave Technol. 2016, 34, 5239–5243. [Google Scholar] [CrossRef]
- Safavieh, M.; Ahmed, M.U.; Tolba, M.; Zourob, M. Microfluidic electrochemical assay for rapid detection and quantification of Escherichia coli. Biosens. Bioelectron. 2012, 31, 523–528. [Google Scholar] [CrossRef] [PubMed]
- Narsaiah, K.; Jha, S.N.; Bhardwaj, R.; Sharma, R.; Kumar, R. Optical biosensors for food quality and safety assurance-a review. J. Food Sci. Technol. 2012, 49, 383–406. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, X.; Lin, Z. Recent developments and applications of surface plasmon resonance biosensors for the detection of mycotoxins in foodstuffs. Food Chem. 2012, 132, 1549–1554. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.S.; Fan, S.K. Microfluidic Surface Plasmon Resonance Sensors: From Principles to Point-of-Care Applications. Sensors 2016, 16, 1175. [Google Scholar] [CrossRef] [PubMed]
- Pennacchio, A.; Ruggiero, G.; Staiano, M.; Piccialli, G.; Oliviero, G.; Lewkowicz, A.; Synak, A.; Bojarski, P.; D’Auriaa, S. A surface plasmon resonance based biochip for the detection of patulin toxin. Opt. Mater. 2014, 36, 1670–1675. [Google Scholar] [CrossRef]
- Zordan, M.D.; Grafton, M.M.G.; Acharya, G.; Reece, L.M.; Cooper, C.L.; Aronson, A.I.; Park, K.; Leary, J.F. Detection of pathogenic E. coli O157:H7 by a hybrid microfluidic SPR and molecular imaging cytometry device. Cytom. Part A 2010, 75, 155–162. [Google Scholar]
- Zordan, M.D.; Grafton, M.M.G.; Acharya, G.; Reece, L.M.; Aronson, A.I.; Park, K.; Leary, J.F. A microfluidic-based hybrid SPR/molecular imaging biosensor for the multiplexed detection of foodborne pathogens. Proc. Spie—Int. Soc. Opt. Eng. 2009, 7167, 1–10. [Google Scholar]
- Tokel, O.; Yildiz, U.H.; Inci, F.; Durmus, N.G.; Ekiz, O.O.; Turker, B.; Cetin, C.; Rao, S.; Sridhar, K.; Natarajan, N.; et al. Portable Microfluidic Integrated Plasmonic Platform for Pathogen Detection. Sci. Rep. 2015, 5, 9152. [Google Scholar] [CrossRef] [PubMed]
- Reig, B.; Bardinal, V.; Camps, T.; Doucet, J.B. A miniaturized VCSEL-based system for optical sensing in a microfluidic channel. Sensors 2012, 23, 1–4. [Google Scholar]
- Ohk, S.H.; Koo, O.K.; Sen, T.; Yamamoto, C.M.; Bhunia, A.K. Antibody–aptamer functionalized fibre-optic biosensor for specific detection of Listeria monocytogenes from food. J. Appl. Microbiol. 2010, 109, 808–817. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Zhang, W.; Wang, Z.; Liu, T.; Zhang, Y. Progress on fiber-optic evanescent wave biosensor technique in food safety detection. J. Food Saf. Qual. 2014, 5, 3971–3974. [Google Scholar]
- Li, M.; Zhao, F.; Zeng, J.; Qi, J.; Lu, J.; Shih, W.C. Microfluidic surface-enhanced Raman scattering sensor with monolithically integrated nanoporous gold disk arrays for rapid and label-free biomolecular detection. J. Biomed. Opt. 2014, 19, 111611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mungroo, N.A.; Oliveira, G.; Neethirajan, S. SERS based point-of-care detection of food-borne pathogens. Microchim. Acta 2016, 183, 697–707. [Google Scholar] [CrossRef]
- Gilli, E. Optical biosensor system with integrated microfluidic sample preparation and TIRF based detection. Proc. Spie—Int. Soc. Opt. Eng. 2013, 8774, 140–144. [Google Scholar]
- Setterington, E.B.; Alocilja, E.C. Electrochemical Biosensor for Rapid and Sensitive Detection of Magnetically Extracted Bacterial Pathogens. Biosensors 2012, 2, 15–31. [Google Scholar] [CrossRef] [Green Version]
- Campuzano, S.; Yanez-Sedeno, P.; Pingarron, J.M. Molecular Biosensors for Electrochemical Detection of Infectious Pathogens in Liquid Biopsies: Current Trends and Challenges. Sensors 2017, 17, 2533. [Google Scholar] [CrossRef]
- Dong, S.; Zhou, J.; Hui, D.; Pang, X.; Wang, Q.; Zhang, S.; Wang, L. Interaction between edge dislocations and amorphous interphase in carbon nanotubes reinforced metal matrix nanocomposites incorporating interface effect. Int. J. Solids Struct. 2014, 51, 1149–1163. [Google Scholar] [CrossRef]
- Ligaj, M.; Tichoniuk, M.; Gwiazdowska, D.; Filipiak, M. Electrochemical DNA biosensor for the detection of pathogenic bacteria Aeromonas hydrophila. Electrochim. Acta 2014, 128, 67–74. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, D.; Cai, G.; Xiong, Y.; Li, Y.; Wang, M.; Hou, H.; Lin, J. Fast and sensitive detection of foodborne pathogen using electrochemical impedance analysis, urease catalysis and microfluidics. Biosens. Bioelectron. 2016, 86, 770–776. [Google Scholar] [CrossRef]
- Liu, H.T.; Wen, Z.Y.; Xu, Y.; Shang, Z.G.; Peng, J.L.; Tian, P. An integrated microfluidic analysis microsystems with bacterial capture enrichment and in-situ impedance detection. Mod. Phys. Lett. B 2017, 31, 196–199. [Google Scholar] [CrossRef]
- Wang, X.; Niessner, R.; Tang, D.; Knopp, D. Nanoparticle-based immunosensors and immunoassays for aflatoxins. Anal. Chim. Acta 2016, 912, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Jing, H.; Cao, X.; Huang, K.; Luo, Y.; Xu, W. Development of a double-antibody sandwich ELISA for rapid detection of Bacillus Cereus in food. Sci. Rep. 2016, 6, 16092. [Google Scholar] [CrossRef] [PubMed]
- Rasooly, A.; Bruck, H.A.; Kostov, Y. An ELISA Lab-on-a-Chip (ELISA-LOC). Humana Press 2013, 949, 451–471. [Google Scholar]
- Thaitrong, N.; Charlermroj, R.; Himananto, O.; Seepiban, C.; Karoonuthaisiri, N. Implementation of microfluidic sandwich ELISA for superior detection of plant pathogens. PLoS ONE 2013, 8, e83231. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.H.; Ma, Y.D.; Chung, Y.D.; Lee, G.B. An integrated microfluidic system for dual aptamer assay utilizing magnetic-composite-membranes. IEEE Int. Conf. Nano/Micro Eng. Mol. Syst. 2017, 4, 438–441. [Google Scholar]
- Yanagisawa, N.; Dutta, D. Enhancement in the Sensitivity of Microfluidic Enzyme-Linked Immunosorbent Assays through Analyte Preconcentration. Anal. Chem. 2012, 84, 7029. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.Q.; Liu, S.L.; Zhao, W.; Zhang, W.P.; Yu, X.; Li, Y.; Li, A.J.; Pang, D.W.; Zhang, Z.L. A Simple Point-of-Care Microfluidic Immunomagnetic Fluorescence Assay for Pathogens. Anal. Chem. 2013, 85, 2645–2651. [Google Scholar] [CrossRef]
- Kanayeva, D.A.; Wang, R.; Rhoads, D.; Erf, G.F.; Slavik, M.F.; Tung, S.; Li, Y. Efficient separation and sensitive detection of Listeria monocytogenes using an impedance immunosensor based on magnetic nanoparticles, a microfluidic chip, and an interdigitated microelectrode. J. Food Prot. 2012, 75, 1951–1959. [Google Scholar] [CrossRef]
- Dector, A.; Galindo-De-La-Rosa, J.; Amaya-Cruz, D.M.; Ortíz-Verdín, A.; Guerra-Balcázar, M.; Olivares-Ramírez, J.M.; Arriaga, L.G.; Ledesma-García, J. Towards autonomous lateral flow assays: Paper-based microfluidic fuel cell inside an HIV-test using a blood sample as fuel. Int. J. Hydrog. Energy 2017, 42, 29–32. [Google Scholar] [CrossRef]
- Hsieh, H.; Dantzler, J.; Weigl, B. Analytical Tools to Improve Optimization Procedures for Lateral Flow Assays. Diagnostics 2017, 7, 29. [Google Scholar] [CrossRef]
- Doller, C.; Jakubik, J. Direct solid-phase radioimmunoassay for the detection of Aujeszky’s disease antibodies. Zent. Bakteriol. A 1980, 247, 1–7. [Google Scholar]
- Yao, L.; Wang, L.; Huang, F.; Cai, G.; Xi, X.; Lin, J. A microfluidic impedance biosensor based on immunomagnetic separation and urease catalysis for continuous-flow detection of E. coli O157:H7. Sens. Actuators B Chem. 2018, 259, 2657. [Google Scholar] [CrossRef]
- Mangal, M.; Bansal, S.; Sharma, S.K.; Gupta, R.K. Molecular Detection of Food Borne Pathogens: A Rapid and Accurate Answer to Food Safety. Crit. Rev. Food Sci. Nutr. 2016, 56, 1568–1584. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, T.H. An automated all-in-one microfludic device for parallel solid phase DNA extraction and droplet-inoil PCR analysis. IEEE Int. Conf. Micro Electro Mech. Syst. 2010, 9, 971–974. [Google Scholar]
- Zhang, C.; Wang, H.; Xing, D. Multichannel oscillatory-flow multiplex PCR microfluidics for high-throughput and fast detection of foodborne bacterial pathogens. Biomed. Microdevices 2011, 13, 885. [Google Scholar] [CrossRef]
- Tourlousse, D.M.; Ahmad, F.; Stedtfeld, R.D.; Seyrig, G.; Tiedje, J.M.; Hashsham, S.A. A polymer microfluidic chip for quantitative detection of multiple water- and foodborne pathogens using real-time fluorogenic loop-mediated isothermal amplification. Biomed. Microdevices 2012, 14, 769–778. [Google Scholar] [CrossRef] [PubMed]
- Uddin, S.M.; Ibrahim, F.; Sayad, A.A.; Thiha, A.; Pei, K.X.; Mohktar, M.S.; Hashim, U.; Cho, J.M.; Thong, K.L. A portable automatic endpoint detection system for amplicons of loop mediated isothermal amplification on microfluidic compact disk platform. Sensors 2015, 15, 5376–5389. [Google Scholar] [CrossRef]
- Sun, Y.; Quyen, T.L.; Hung, T.Q.; Chin, W.H.; Wolff, A.; Bang, D.D. A lab-on-a-chip system with integrated sample preparation and loop-mediated isothermal amplification for rapid and quantitative detection of Salmonella spp. in food samples. Lab Chip 2015, 15, 1898–1904. [Google Scholar] [CrossRef] [Green Version]
- Lutz, S.; Weber, P.; Focke, M.; Faltin, B.; Hoffmann, J.; Müller, C.; Mark, D.; Roth, G.; Munday, P.; Armes, N.; et al. Microfluidic lab-on-a-foil for nucleic acid analysis based on isothermal recombinase polymerase amplification (RPA). Lab Chip 2010, 10, 887–893. [Google Scholar] [CrossRef]
- Tortajada-Genaro, L.A.; Santiago-Felipe, S.; Amasia, M.; Russom, A.; Maquieira, Á. Isothermal solid-phase recombinase polymerase amplification on microfluidic digital versatile discs (DVDs). RSC Adv. 2015, 5, 29987–29995. [Google Scholar] [CrossRef] [Green Version]
- Mauk, M.G.; Liu, C.; Song, J.; Bau, H.H. Integrated Microfluidic Nucleic Acid Isolation, Isothermal Amplification, and Amplicon Quantification. Microarrays 2015, 4, 474–489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, B.; Fu, K.; Liu, Y.; Ding, X.; Hu, J.; Wu, W.; Xu, K.; Song, X.L.; Wang, J.; Mu, Y.; et al. Development of a self-priming PDMS/paper hybrid microfluidic chip using mixed-dye-loaded loop-mediated isothermal amplification assay for multiplex foodborne pathogens detection. Anal. Chim. Acta 2018, 1040, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.L.; Zhao, X.H. Detection of viable but non-culturable Escherichia coli O157:H7 by PCR in combination with propidium monoazide. 3 Biotech 2017, 8, 28. [Google Scholar] [CrossRef] [PubMed]
- Li, M.F.; Li, L.M.; Liu, R.Y. Application of paper based microfluidic chip technology in food safety detection. J. Food Saf. Qual. 2018, 38, e12412. [Google Scholar]





| Material Type | Classification | Representative | Methods of Preparation | Advantages | Disadvantages | Application | References |
|---|---|---|---|---|---|---|---|
| organic material | ------- | glass/quartz | photolithography and etching techniques | cheap and easy to obtain, reusable, good light transmission and electroosmosis, good electrical insulation and corrosion resistance | complex manufacturing process, time-consuming and high cost, fragile | gas chromatography and capillary electrophoresis (CE) and electrochemical detection, organic synthesis and droplet formation, PCR | [41,42] |
| silicon material | silicon/silicon dioxide | etching techniques | mature process, good thermal stability and inertness. | high cost of materials, opaque, brittle, poor electrical insulation, and low adhesion coefficient | organic synthesis and droplet formation, PCR and CE | [43,44] | |
| elastomers | polydimethylsiloxane (PDMS) | molding and soft lithography | Low cost and easy to use, non-toxic and transparent, excellent chemical inertness and light transmission | Incompatibility of organic solvents and poor pressure resistance, low thermal conductivity and immature processing technology | protein crystallization and bioculture, PCR | [45,46] | |
| Polymer materials | thermosets | SU-8 photoresist and polyimide | photopolymerization and casting | High resistance of temperature and most solvents, transparent and reusable | high cost of materials | CE, organic synthesis and droplet formation, PCR | [47,48] |
| thermoplastics | poly (methyl methacrylate (PMMA) polystyrene (PS) and polycarbonate (PC) | hot embossing and laser ablation | good electrical insulation and light transmission, low cost and easy to use, simple preparation and high precision | Non-breathable, high-cost preparation equipment and rough process | CE and PCR, droplet formation | [49,50] | |
| perfluoropolymers | perfluoroalkoxy (PFA) and fluorinated ethylene propylene | photolithography | Good inertness and antifouling properties, transparent and soft | poor adhesion | environmental monitoring and food analysis | [51] | |
| Special materials | hydrogels | polyvinyl alcohol (PVA) | photopolymerization, casting | high permeability and controllable aperture, allowing small molecules or even biological particles to diffuse, and biocompatible | difficult to store | 3D bioculture | [52] |
| ceramics | polysiloxane | soft lithography and laser ablation | high resistance of temperature and pressure | poor light transmission, fragile | suitable for applications under harsh conditions | [53] | |
| paper | analysis filter paper | photolithography and printing | high permeability and low cost, portable and easy to use | easy to damage and disposable | bioculture | [54] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, X.; Li, M.; Liu, Y. Microfluidic-Based Approaches for Foodborne Pathogen Detection. Microorganisms 2019, 7, 381. https://doi.org/10.3390/microorganisms7100381
Zhao X, Li M, Liu Y. Microfluidic-Based Approaches for Foodborne Pathogen Detection. Microorganisms. 2019; 7(10):381. https://doi.org/10.3390/microorganisms7100381
Chicago/Turabian StyleZhao, Xihong, Mei Li, and Yao Liu. 2019. "Microfluidic-Based Approaches for Foodborne Pathogen Detection" Microorganisms 7, no. 10: 381. https://doi.org/10.3390/microorganisms7100381