Molecular Diversity and Biochemical Content in Two Invasive Alien Species: Looking for Chemical Similarities and Bioactivities
Abstract
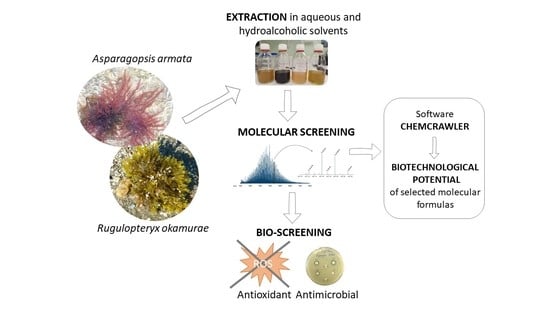
:1. Introduction
2. Results
2.1. Biochemical Composition
2.2. Molecular Diversity and Published Bioactivities
| Abbreviation | Formulas | Top Isomer(s) | Molecule Iype | Bio-Activities | Ref. |
|---|---|---|---|---|---|
| dH2O | C18H34O5 | Pinellic acid | Trihydroxyoctadecenoic acid | Adjuvant and an anti-inflammatory agent | [53] |
| 9,10,13-TriHOME | Monounsaturated fatty acid | It has a role as a human blood serum metabolite | [91] | ||
| dH2O:EtOH(1:1) dH2O:EtOH(1:4) dH2O:MeOH(1:1) | C18H32O16 | Maltotriose | Trisaccharide | Sweetener produced from starch Used in baked goods, and beer and other fermented drinks production Prebiotic effect | [61,62,63] |
| Dextran | Glucan | Anti-thrombotic effect, blood viscosity reducer and a volume expander Bread texture improvers | [64,65] | ||
| dH2O:EtOH(1:1) dH2O:EtOH(1:4) dH2O:MeOH(1:1) | C10H18N2O3 | Dethiobiotin | Conjugate acid | Agonists of nuclear receptor subfamily 2, group E, member 3 (NR2E3) | [92] |
| dH2O:EtOH(1:1) | C8H12N2O2 | Pyridoxamine | Monohydroxypyridine | Human, Saccharomyces cerevisiae, Escherichia coli, plant, and mouse metabolite, an iron chelator, and a nephroprotective agent | [93] |
| dH2O:EtOH(1:1) | C7H6O5 | Gallic acid | Phenolic | Astringent, a cyclooxygenase 2 inhibitor, a plant metabolite, antioxidant, antineoplastic agent, a human xenobiotic metabolite, an EC 1.13.11.33 (arachidonate 15-lipoxygenase) inhibitor, an apoptosis inducer, and a geroprotector | [59] |
| dH2O:MeOH(1:1) | C6H13N3O3 | Citrulline | Non-essential amino acid | EC 1.14.13.39 (nitric oxide synthase) inhibitor, a protective agent, a nutraceutical, a micronutrient, and a human, an Escherichia coli, a Saccharomyces cerevisiae, and a mouse metabolite | [60] |
| dH2O:MeOH(1:4) | C22H28O11 | Prim-O-glucosylcimifugin | Chromone | Anti-pyretic, analgesic and anti-inflammatory activities Used in many Kampo prescriptions Anti-rheumatoid, anti-inflammatory, immunosuppressive, and pain-relieving properties | [52,55,56,58,94,95,96] |
| 4′-O-Glucosyl-5-O-methylvisamminol | Chromone | Analgesic, anti-inflammatory, anti-psoriasis, and antiplatelet aggregation effects | [54,57] | ||
| dH2O:MeOH(1:4) | C23H30O11 | Yadanzioside D | Anti-Tobacco Mosaic Virus (TMV) activity | [67,97] | |
| dH2O:MeOH(1:4) | C20H24O8 | Vernodalol | Sesquiterpene lactone | Angeloylated germacranolides from Daucus virgatus and their plasmodium transmission blocking activity | [66] |
| dH2O:MeOH(1:4) | C6H8O6 | Ascorbic acid (Vitamin C) | Natural water-soluble vitamin | Potent reducing and antioxidant agent found in citrus and other fruits, and in vegetables | [68] |
| Abbreviation | Formulas | Top Isomer(s) | Molecule Type | Bio-Activities | Refer. |
|---|---|---|---|---|---|
| dH2O | C10H8O5 | Fraxetin | Hydroxycoumarin | Antimicrobial agent, apoptosis inhibitor, apoptosis inducer, antioxidant, anti-inflammatory agent, a hepatoprotective agent, antibacterial agent, and a hypoglycemic agent | [71,72,73] |
| dH2O | C20H34O5 | Prostaglandin F2alpha | Prostaglandin | Oxytocic, luteolytic, abortifacient, and vasocontractile activities | [69] |
| Alprostadil | Prostaglandin | Potent vasodilator agent that increases peripheral blood flow, inhibits platelet aggregation, and induces bronchodilation | [70] | ||
| dH2O | C18H28O3 | 12-Oxo-phytodienoic acid | Unsaturated fatty acid | Inhibitor of the protein serine/threonine kinase 33 (STK33) | [98] |
| dH2O | C20H34O6 | Thromboxane B2 | Stable metabolite | Human and mouse metabolite | [99] |
| 6-Keto-prostaglandin F1alpha | Prostaglandin | Human and mouse metabolite | [100] | ||
| dH2O:EtOH(1:1) dH2O:MeOH(1:1) dH2O:MeOH(1:4) | C20H24O8 | Vernodalol | Sesquiterpene lactone | Angeloylated germacranolides from Daucus virgatus and their plasmodium transmission blocking activity | [66] |
| dH2O:EtOH(1:1) dH2O:EtOH(1:4) dH2O:EtOH(1:4) dH2O:MeOH(1:4) | C17H22O4 | 1-Dehydro-[6]-gingerdione | Hydroxycinnamic acid | Antiallergic potential | [101] |
| dH2O:EtOH(1:1) dH2O:MeOH(1:1) dH2O:MeOH(1:4) | C14H16O5 | 1′-Acetoxyeugenol acetate | Phenylpropanoid | Anti-breast cancer properties | [102] |
| dH2O:EtOH(1:1) dH2O:MeOH(1:1) | C12H14O2 | Precocene I | Chromene, aromatic ether | Member of precocenes and a plant metabolite | [103] |
| dH2O:EtOH(1:1) dH2O:MeOH(1:4) | C12H14O3 | Acetyleugenol | Phenol, benzoate ester | Antivirulence potential against pathogenic bacteria of medical importance | [104] |
| dH2O:EtOH(1:1) dH2O:MeOH(1:1) dH2O:MeOH(1:4) | C24H34O10 | 3. ′-Hydroxy-T2 Toxin | Sesquiterpene | A trichothecene with mycotoxin effects | [79,105] |
| dH2O:EtOH(1:1) dH2O:MeOH(1:4) | C19H28O6 | Tirotundin | Sesquiterpene lactone | Antidiabetic effect through PPARγ pathway | [106] |
| dH2O:EtOH(1:1) dH2O:MeOH(1:1) dH2O:EtOH(1:4) dH2O:MeOH(1:4) | C19H24O3 | 2-Methoxyestrone | Steroid | Indicator of the risk of prostate, colorectal, and breast cancer | [107,108,109] |
| dH2O:EtOH(1:1) dH2O:MeOH(1:1) dH2O:EtOH(1:4) dH2O:MeOH(1:4) | C15H18O4 | Helenalin | Sesquiterpene lactone | Anti-inflammatory and anti-neoplastic agent Anti-tumoral effects | [75,110,111] |
| Parthenin | Sesquiterpene lactone | It is genotoxic, allergenic, and an irritant It is believed to be responsible for the dermatitis caused by Parthenium hysterophorus | [77,112] | ||
| dH2O:EtOH(1:1) dH2O:MeOH(1:1) dH2O:EtOH(1:4) dH2O:MeOH(1:4) | C15H16O3 | Osthole | Coumarin | Antioxidant and immunomodulatory properties, commonly applied in clinical practice of Traditional Chinese Medicine for cancer, inflammation, etc. It has estrogen-like effects that prevent osteoporosis and reduce bone loss in ovariectomized rats by activation of β-catenin-BMP signaling It is a chromatin regulator implicated in the inhibition of histone deacetylases (HDACs) in order to cure or prevent cancer | [80,81,82,113] |
| Batatasin III | Stilbenoid | Anti-inflammatory and anti-cancer activity | [114,115] | ||
| dH2O:EtOH(1:1) dH2O:EtOH(1:4) dH2O:MeOH(1:1) dH2O:MeOH(1:4) | C15H18O3 | Santonin | Propionic acid | Effective for treating intestinal roundworms | [116,117] |
| Irofulven | Sesquiterpene | A natural toxin with anti-cancer potential isolated from the fungus Omphalotus illudens | [59] | ||
| Xanthatin | Sesquiterpene lactone | Tumor suppressor function | [78] | ||
| dH2O:EtOH(1:1) dH2O:EtOH(1:4) dH2O:MeOH(1:1) dH2O:MeOH(1:4) | C17H20O7 | Dihydroscandenolide | Sesquiterpene lactone | Anti-fungal and anti-bacterial activity | [74] |
| Yomogiartemin | Sesquiterpene lactone | Anti-malarial activity | [76] | ||
| dH2O:EtOH(1:4) dH2O:MeOH(1:1) | C23H34O7 | Nargenicin A1 | Saturated alicyclic polyketide | Bacterial macrolide Anti-cancer activity, immunomodulation, and cell protective effect | [118] |
| dH2O:EtOH(1:1) dH2O:EtOH(1:4) dH2O:MeOH(1:1) dH2O:MeOH(1:4) | C18H24O5 | Zearalanone, β-,α-Zearalenol | Nonsteroidal estrogen | Mycotoxin It has major effects on reproduction in females, it causes cytotoxicity, neurotoxicity, and oxidative stress at the molecular level | [119,120] |
| Formulas | Top Isomer(s) | Bio-Activities | References |
|---|---|---|---|
| C6H6O3 | Maltol | An antioxidant found in Korean red ginseng | [85] |
| Pyrogallol | Highly cytotoxic effect on human lung cancer cell lines, induces apoptosis in endothelial cells. Antibacterial activity against Staphylococcus epidermidis and Staphylococcus aureus | [84,87,88] | |
| C8H8O2 | Phenylacetic acid | Antimetabolite useful in cancer chemotherapy | [121] |
| C6H6O4 | Kolic acid | NF-kappaB inhibitor, skin lightening agent, an EC 1.10.3.1 (catechol oxidase), EC 1.10.3.2 (laccase), EC 1.13.11.24 (quercetin 2,3-dioxygenase), EC 1.14.18.1 (tyrosinase), and EC 1.4.3.3 (D-amino-acid oxidase) inhibitor | [122] |
| C10H10O4 | Ferulic acid | Antioxidant, a MALDI matrix material, a plant metabolite, an anti-inflammatory agent, an apoptosis inhibitor, and a cardioprotective agent | [123] |
| C11H10O4 | Scoparone | Major constituent of the Chinese herbal medicine Yin Chen Hao, exhibits anti-inflammatory, anti-allergic, and anti-tumor activities | [124] |
| C10H8O5 | Fraxetin | An Arabidopsis thaliana metabolite, antimicrobial agent, apoptosis inhibitor and inducer, antioxidant, anti-inflammatory, hepatoprotective agent, antibacterial, and hypoglycemic agent | [125] |
| C12H12O4 | Hispolon | Anti-cancer activity | [83] |
| Eugenitin | Anti-leishmanial and cytotoxicity assays | [89] | |
| C14H12O3 | Resveratrol | A phytoalexin, antioxidant, glioma-associated oncogene inhibitor, and geroprotector found in high concentrations in red grapes | [86] |
| C14H14O4 | Marmesin | Potential natural UV-A-filtering product | [90] |
2.3. Antioxidant Capacity
2.4. Antimicrobial Activity
2.5. Correlations and Principal Component Analysis
3. Discussion
4. Materials and Methods
4.1. Algal Material
4.2. Preparation of Algal Extracts
4.3. Biochemical Composition
4.3.1. Internal Carbon, Nitrogen, and Sulphur
4.3.2. Ashes
4.3.3. Total Proteins
4.3.4. Carbohydrates
4.3.5. Lipids
4.3.6. Phenolic Compounds
4.4. Molecular Diversity and Published Bioactivities
4.5. Antioxidant Capacity (ABTS and DPPH)
4.6. Antimicrobial Activity
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Schaffelke, B.; Smith, J.E.; Hewitt, C.L. Introduced Macroalgae—A Growing Concern. J. Appl. Phycol. 2006, 18, 529–541. [Google Scholar] [CrossRef]
- Molnar, J.L.; Gamboa, R.L.; Revenga, C.; Spalding, M.D. Assessing the global threat of invasive species to marine biodiversity. Front. Ecol. Environ. 2008, 6, 485–492. [Google Scholar] [CrossRef]
- Raffo, M.P.; Eyras, M.C.; Iribarne, O.O. The invasion of Undaria pinnatifida to a Macrocystis pyrifera kelp in patagonia (Argentina, south-west Atlantic). J. Mar. Biol. Assoc. United Kingd. 2009, 89, 1571–1580. [Google Scholar] [CrossRef]
- Galil, B.S.; Marchini, A.; Occhipinti-Ambrogi, A. East is east and West is west? Management of marine bioinvasions in the Mediterranean Sea. Estuar. Coast. Shelf Sci. 2018, 201, 7–16. [Google Scholar] [CrossRef]
- Viard, F.; Comtet, T. Applications of DNA-based methods for the study of biological invasions. In Biological Invasions in Changing Ecosystems: Vectors, Ecological Impacts, Management and Predictions; De Gruyter: Berlin, Germany, 2015; pp. 411–435. ISBN 9783110438666. [Google Scholar]
- Sorte, C.J.B.; Williams, S.L.; Zerebecki, R.A. Ocean warming increases threat of invasive species in a marine fouling community. Ecology 2010, 91, 2198–2204. [Google Scholar] [CrossRef] [PubMed]
- Pimentel, D.; Zuniga, R.; Morrison, D. Update on the environmental and economic costs associated with alien-invasive species in the United States. Ecol. Econ. 2005, 52, 273–288. [Google Scholar] [CrossRef]
- Xu, H.; Ding, H.; Li, M.; Qiang, S.; Guo, J.; Han, Z.; Huang, Z.; Sun, H.; He, S.; Wu, H.; et al. The distribution and economic losses of alien species invasion to China. Biol. Invasions 2006, 8, 1495–1500. [Google Scholar] [CrossRef]
- Vilà, M.; Basnou, C.; Pyšek, P.; Josefsson, M.; Genovesi, P.; Gollasch, S.; Nentwig, W.; Olenin, S.; Roques, A.; Roy, D.; et al. How well do we understand the impacts of alien species on ecosystem services? A pan-European, cross-taxa assessment. Front. Ecol. Environ. 2010, 8, 135–144. [Google Scholar] [CrossRef] [Green Version]
- Zenetos, A.; Gofas, S.; Verlaque, M.; Cinar, M.E.; Garcia Raso, J.E.; Bianchi, C.N.; Morri, C.; Azzurro, E.; Bilecenoglu, M.; Froglia, C.; et al. Alien species in the Mediterranean Sea by 2010. A contribution to the application of European Union’s Marine Strategy Framework Directive (MSFD). Part I. Spatial distribution. Mediterr. Mar. Sci. 2010, 11, 381. [Google Scholar] [CrossRef] [Green Version]
- Zenetos, A.; Gofas, S.; Morri, C.; Rosso, A.; Violanti, D.; Garcia Raso, J.E.; Cinar, M.E.; Almogi-Labin, A.; Ates, A.S.; Azzurro, E.; et al. Alien species in the Mediterranean Sea by 2012. A contribution to the application of European Union’s Marine Strategy Framework Directive (MSFD). Part 2. Introduction trends and pathways. Mediterr. Mar. Sci. 2012, 13, 328. [Google Scholar] [CrossRef]
- Kraan, S. Undaria marching on; late arrival in the Republic of Ireland. J. Appl. Phycol. 2017, 29, 1107–1114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bunicontro, M.P.; Marcomini, S.C.; Casas, G.N. Environmental Impacts of an Alien Kelp Species (Undaria pinnatifida, Laminariales) Along the Patagonian Coasts. In Coastal Research Library; Springer: Berlin/Heidelberg, Germany, 2019; Volume 29, pp. 373–396. [Google Scholar]
- Qi, L.; Hu, C.; Xing, Q.; Shang, S. Long-term trend of Ulva prolifera blooms in the western Yellow Sea. Harmful Algae 2016, 58, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Cope, R.C.; Ross, J.V.; Wittmann, T.A.; Watts, M.J.; Cassey, P. Predicting the Risk of Biological Invasions Using Environmental Similarity and Transport Network Connectedness. Risk Anal. 2019, 39, 35–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sardain, A.; Sardain, E.; Leung, B. Global forecasts of shipping traffic and biological invasions to 2050. Nat. Sustain. 2019, 2, 274–282. [Google Scholar] [CrossRef]
- Zhang, J.; Shi, J.; Gao, S.; Huo, Y.; Cui, J.; Shen, H.; Liu, G.; He, P. Annual patterns of macroalgal blooms in the Yellow Sea during 2007–2017. PLoS ONE 2019, 14, e0210460. [Google Scholar] [CrossRef] [Green Version]
- Pacheco, D.; Araújo, G.S.; Cotas, J.; Gaspar, R.; Neto, J.M.; Pereira, L. Invasive Seaweeds in the Iberian Peninsula: A Contribution for Food Supply. Mar. Drugs 2020, 18, 560. [Google Scholar] [CrossRef]
- Ministerio Para la Transición Ecológica y el Reto Demográfico. Catálogo Español de Especies Exóticas Invasorasicono Barra Herramientas. Available online: https://www.miteco.gob.es/es/biodiversidad/temas/c (accessed on 25 November 2022).
- Horridge, G.A. Occurrence of asparagopsis armata harv. on the scilly Isles. Nature 1951, 167, 732–733. [Google Scholar] [CrossRef]
- Streftaris, N.; Zenetos, A. Alien marine species in the Mediterranean - the 100 ‘worst invasives’ and their impact. Mediterr. Mar. Sci. 2006, 7, 87–118. [Google Scholar] [CrossRef] [Green Version]
- Zanolla, M.; Carmona, R.; Mata, L.; De la Rosa, J.; Sherwood, A.; Barranco, C.N.; Muñoz, A.R.; Altamirano, M. Concise review of the genus Asparagopsis Montagne, 1840. J. Appl. Phycol. 2022, 34, 1–17. [Google Scholar] [CrossRef]
- Feldmann, J.; Feldmann, G. Recherches sur les Bonnemaisoniacées et leur alternance de générations. Ann. Sci. Nat. Bot. Biol. Veg. 1942, 3, 75–175. [Google Scholar]
- El Aamri, F.; Idhalla, M.; Tamsouri, M.N. Occurrence of the invasive brown seaweed Rugulopteryx okamurae (E.Y. Dawson) I.K. Hwang, W.J. Lee & H.S. Kim (Dictyotales, Phaeophyta) in Morocco (Mediterranean Sea). Mediterr. Fish. Aquac. Res. 2018, 1, 92–96. [Google Scholar]
- Altamirano Jeschke, M.; De la Rosa Álamos, J.; Martínez Medina, F.J. Arribazones de la especia exótica Rugulopteryx okamurae (E.Y.Dawson) en el Estrecho de Gibraltar. 2016. Available online: https://riuma.uma.es/xmlui/handle/10630/12433 (accessed on 10 September 2022).
- Ocaña, O.; Alfonso-Carrillo, J.; Ballesteros, E. Massive proliferation of a dictyotalean species (Phaeophyceae, Ochrophyta) through the Strait of Gibraltar (Research note). Rev. la Acad. Canar. Cienc. 2016, 28, 165–170. [Google Scholar]
- García-Gómez, J.C.; Florido, M.; Olaya-Ponzone, L.; Rey Díaz de Rada, J.; Donázar-Aramendía, I.; Chacón, M.; Quintero, J.J.; Magariño, S.; Megina, C. Monitoring Extreme Impacts of Rugulopteryx okamurae (Dictyotales, Ochrophyta) in El Estrecho Natural Park (Biosphere Reserve). Showing Radical Changes in the Underwater Seascape. Front. Ecol. Evol. 2021, 9, 639161. [Google Scholar] [CrossRef]
- García-Gómez, J.C.; Sempere-Valverde, J.; González, A.R.; Martínez-Chacón, M.; Olaya-Ponzone, L.; Sánchez-Moyano, E.; Ostalé-Valriberas, E.; Megina, C. From exotic to invasive in record time: The extreme impact of Rugulopteryx okamurae (Dictyotales, Ochrophyta) in the strait of Gibraltar. Sci. Total Environ. 2020, 704, 135408. [Google Scholar] [CrossRef] [PubMed]
- Sempere-Valverde, J.; Ostalé-Valriberas, E.; Maestre, M.; González Aranda, R.; Bazairi, H.; Espinosa, F. Impacts of the non-indigenous seaweed Rugulopteryx okamurae on a Mediterranean coralligenous community (Strait of Gibraltar): The role of long-term monitoring. Ecol. Indic. 2021, 121, 107135. [Google Scholar] [CrossRef]
- García-Gómez, J.C.; Florido, M.; Olaya-Ponzone, L.; Sempere-Valverde, J.; Megina, C. The Invasive Macroalga Rugulopteryx okamurae: Substrata Plasticity and Spatial Colonization Pressure on Resident Macroalgae. Front. Ecol. Evol. 2021, 9, 259. [Google Scholar] [CrossRef]
- Bermejo, R.; Vergara, J.J.; Hernández, I. Application and reassessment of the reduced species list index for macroalgae to assess the ecological status under the Water Framework Directive in the Atlantic coast of Southern Spain. Ecol. Indic. 2012, 12, 46–57. [Google Scholar] [CrossRef]
- Bermejo, R.; Mangialajo, L.; Vergara, J.J.; Hernández, I. Comparison of two indices based on macrophyte assemblages to assess the ecological status of coastal waters in the transition between the Atlantic and Mediterranean eco-regions. J. Appl. Phycol. 2014, 26, 1899–1909. [Google Scholar] [CrossRef]
- Figueroa, F.L.; Flores-Moya, A.; Vegara, J.J.; Korbee, N.; Hernández, I. Autochtonous seaweeds. In The Mediterranean Sea: Its History and Present Challenges; Springer: Dordrecht, The Netherlands, 2012. [Google Scholar]
- Verlaque, M.; Steen, F.; De Clerck, O. Rugulopteryx (Dictyotales, Phaeophyceae), a genus recently introduced to the Mediterranean. Phycologia 2009, 48, 536–542. [Google Scholar] [CrossRef]
- Pinteus, S.; Lemos, M.F.L.; Alves, C.; Neugebauer, A.; Silva, J.; Thomas, O.P.; Botana, L.M.; Gaspar, H.; Pedrosa, R. Marine invasive macroalgae: Turning a real threat into a major opportunity - the biotechnological potential of Sargassum muticum and Asparagopsis armata. Algal Res. 2018, 34, 217–234. [Google Scholar] [CrossRef]
- Pereira, A.G.; Fraga-Corral, M.; Garcia-Oliveira, P.; Lourenço-Lopes, C.; Carpena, M.; Prieto, M.A.; Simal-Gandara, J. The Use of Invasive Algae Species as a Source of Secondary Metabolites and Biological Activities: Spain as Case-Study. Mar. Drugs 2021, 19, 178. [Google Scholar] [CrossRef] [PubMed]
- Zubia, M.; Fabre, M.S.; Kerjean, V.; Deslandes, E. Antioxidant and cytotoxic activities of some red algae (Rhodophyta) from Brittany coasts (France). Bot. Mar. 2009, 52, 268–277. [Google Scholar] [CrossRef] [Green Version]
- Pinteus, S.; Silva, J.; Alves, C.; Horta, A.; Fino, N.; Rodrigues, A.I.; Mendes, S.; Pedrosa, R. Cytoprotective effect of seaweeds with high antioxidant activity from the Peniche coast (Portugal). Food Chem. 2017, 218, 591–599. [Google Scholar] [CrossRef] [PubMed]
- Pinteus, S.; Rodrigues, A.; Silva, J.; Lokman, C.; Lemos, M.; Pedrosa, R. The marine invasive Asparagopsis armata (Harvey, 1855) as source of bioactive valuable compounds - Antioxidant potential enrichment by Vacuum liquid Chromatography. Front. Mar. Sci. 2016, 3. [Google Scholar] [CrossRef]
- Pinteus, S.; Lemos, M.F.L.; Alves, C.; Silva, J.; Pedrosa, R. The marine invasive seaweeds Asparagopsis armata and Sargassum muticum as targets for greener antifouling solutions. Sci. Total Environ. 2021, 750, 141372. [Google Scholar] [CrossRef]
- Paul, N.A.; De Nys, R.; Steinberg, P.D. Chemical defence against bacteria in the red alga Asparagopsis armata: Linking structure with function. Mar. Ecol. Prog. Ser. 2006, 306, 87–101. [Google Scholar] [CrossRef] [Green Version]
- Salvador, N.; Gómez Garreta, A.; Lavelli, L.; Ribera, M.A. Antimicrobial activity of Iberian macroalgae. Sci. Mar. 2007, 71, 101–113. [Google Scholar] [CrossRef] [Green Version]
- Oumaskour, K.; Boujaber, N.; Etahiri, S.; Assobhei, O. Anti-inflammatory and antimicrobial activities of twenty-tree marine red algae from the coast of Sidi Bouzid (El Jadida-Morocco). Int. J. Pharm. Pharm. Sci. 2013, 5, 145–149. [Google Scholar]
- Rhimou, B.; Hassane, R.; Nathalie, B. Antiviral activity of the extracts of Rhodophyceae from Morocco. Afr. J. Biotechnol. 2010, 9, 7968–7975. [Google Scholar] [CrossRef] [Green Version]
- Ochi, M.; Masui, N.; Kotsuki, H.; Miura, I.; Tokoroyama, T. The structures of fukurinolal and fukurinal, two new diterpenoids from the brown seaweed Dilophus okamurai Dawson. Chem. Lett. 1982, 11, 1927–1930. [Google Scholar] [CrossRef] [Green Version]
- Kurata, K.; Shiraishi, K.; Takato, T.; Taniguchi, K.; Suzuki, M. A New Feeding-Deterrent Diterpenoid from the Brown Alga Dilophus okamurai Dawson. Chem. Lett. 1988, 17, 1629–1632. [Google Scholar] [CrossRef]
- Yamase, H.; Umemoto, K.; Ooi, T.; Kusumi, T. Structures and absolute stereochemistry of five new secospatanes and a spatane isolated from the brown alga Dilophus okamurai Dawson. Chem. Pharm. Bull. 1999, 47, 813–818. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, M.; Yamada, H.; Kurata, K. Dictyterpenoids A and B, two novel diterpenoids with feeding-deterrent activity from the brown alga Dilophus okamurae. J. Nat. Prod. 2002, 65, 121–125. [Google Scholar] [CrossRef]
- De Paula, J.C.; Vallim, M.A.; Teixeira, V.L. What are and where are the bioactive terpenoids metabolites from Dictyotaceae (Phaeophyceae). Rev. Bras. Farmacogn. 2011, 21, 216–228. [Google Scholar] [CrossRef]
- Cuevas, B.; Arroba, A.I.; de los Reyes, C.; Gómez-Jaramillo, L.; González-Montelongo, M.C.; Zubía, E. Diterpenoids from the brown alga Rugulopteryx okamurae and their anti-inflammatory activity. Mar. Drugs 2021, 19, 677. [Google Scholar] [CrossRef] [PubMed]
- Casal-Porras, I.; Zubía, E.; Brun, F.G. Dilkamural: A novel chemical weapon involved in the invasive capacity of the alga Rugulopteryx okamurae in the Strait of Gibraltar. Estuar. Coast. Shelf Sci. 2021, 257, 107398. [Google Scholar] [CrossRef]
- Okuyama, E.; Hasegawa, T.; Matsushita, T.; Fujimoto, H.; Ishibashi, M.; Yamazaki, M. Analgesic components of Saposhnikovia root (Saposhnikovia divaricata). Chem. Pharm. Bull. 2001, 49, 154–160. [Google Scholar] [CrossRef] [Green Version]
- National Center for Biotechnology Information. PubChem Compound Summary for CID 9858729, Pinellic Acid. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/9858729 (accessed on 24 August 2022).
- Fu, J.; Zeng, Z.; Zhang, L.; Wang, Y.; Li, P. 4’-o-b-d-glucosyl-5-o-methylvisamminol ameliorates imiquimod-induced psoriasis-like dermatitis and inhibits inflammatory cytokines production by suppressing the nf-kb and mapk signaling pathways. Braz. J. Med. Biol. Res. 2020, 53, 1–10. [Google Scholar] [CrossRef]
- Baba, K.; Tabata, Y.; Kozawa, M.; Kimura, Y.; Arichi, S. Studies on Chinese traditional medicine Fang-feng (I). Structures and physiological activities of polyacetylene compounds from Saposhnikoviae radix. Shoyakugaku Zasshi 1987, 41, 189–194. [Google Scholar]
- Fuchino, H.; Murase, S.; Hishida, A.; Kawahara, N. Simultaneous UHPLC/MS quantitative analysis and comparison of Saposhnikoviae radix constituents in cultivated, wild and commercial products. J. Nat. Med. 2021, 75, 499–519. [Google Scholar] [CrossRef]
- Kreiner, J.; Pang, E.; Lenon, G.B.; Yang, A.W.H. Saposhnikoviae divaricata: A phytochemical, pharmacological, and pharmacokinetic review. Chin. J. Nat. Med. 2017, 15, 255–264. [Google Scholar] [CrossRef]
- Meng, Y.; Yi, L.; Chen, L.; Hao, J.; Li, D.X.; Xue, J.; Xu, N.Y.; Zhang, Z.Q. Purification, structure characterization and antioxidant activity of polysaccharides from Saposhnikovia divaricata. Chin. J. Nat. Med. 2019, 17, 792–800. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information. PubChem Compound Summary for CID 370, Gallic Acid. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Gallic-a (accessed on 24 August 2022).
- National Center for Biotechnology Information. PubChem Compound Summary for CID 9750, Citrulline. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Citrulli (accessed on 24 August 2022).
- Grijalva-Vallejos, N.; Krogerus, K.; Nikulin, J.; Magalhães, F.; Aranda, A.; Matallana, E.; Gibson, B. Potential application of yeasts from Ecuadorian chichas in controlled beer and chicha production. Food Microbiol. 2021, 98, 103644. [Google Scholar] [CrossRef] [PubMed]
- Jang, E.Y.; Hong, K.B.; Chang, Y.B.; Shin, J.; Jung, E.Y.; Jo, K.; Suh, H.J. In Vitro Prebiotic Effects of Malto-Oligosaccharides Containing Water-Soluble Dietary Fiber. Molecules 2020, 25, 5201. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information. PubChem Compound Summary for CID 192826, alpha-Maltotriose. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/alpha-Ma (accessed on 24 August 2022).
- Lewis, S.M.; Bucher, L.; Heitkemper, M.M.; Harding, M.; Kwong, M.; Roberts, D. Medical-Surgical Nursing: Assessment and Management of Clinical Problems; Elsevier H: Amsterdam, The Netherlands, 2016; ISBN 9780323091473. [Google Scholar]
- Perri, G.; Coda, R.; Rizzello, C.G.; Celano, G.; Ampollini, M.; Gobbetti, M.; De Angelis, M.; Calasso, M. Sourdough fermentation of whole and sprouted lentil flours: In situ formation of dextran and effects on the nutritional, texture and sensory characteristics of white bread. Food Chem. 2021, 355, 129638. [Google Scholar] [CrossRef]
- Sirignano, C.; Snene, A.; Rigano, D.; Tapanelli, S.; Formisano, C.; Luciano, P.; El Mokni, R.; Hammami, S.; Tenoh, A.R.; Habluetzel, A.; et al. Angeloylated Germacranolides from Daucus virgatus and Their Plasmodium Transmission Blocking Activity. J. Nat. Prod. 2017, 80, 2787–2794. [Google Scholar] [CrossRef]
- Yan, X.H.; Chen, J.; Di, Y.T.; Fang, X.; Dong, J.H.; Sang, P.; Wang, Y.H.U.; He, H.P.; Zhang, Z.K.; Hao, X.J. Anti-tobacco mosaic virus (TMV) quassinoids from brucea javanlca (L.) merr. J. Agric. Food Chem. 2010, 58, 1572–1577. [Google Scholar] [CrossRef]
- Vineetha, R.C.; Mathews, V.V.; Nair, R.H. Ascorbic acid and the mitochondria. In Mitochondrial Physiology and Vegetal Molecules: Therapeutic Potential of Natural Compounds on Mitochondrial Health; Elsevier: Amsterdam, The Netherlands, 2021; pp. 613–624. ISBN 9780128215623. [Google Scholar]
- National Center for Biotechnology Information. PubChem Compound Summary for CID 44208919, PGF2alpha. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/prostagl (accessed on 20 August 2022).
- National Center for Biotechnology Information. PubChem Compound Summary for CID 5280723, Alprostadil. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/5280723 (accessed on 20 August 2022).
- Balaha, M.; Ahmed, N.; Geddawy, A.; Kandeel, S. Fraxetin prevented sodium fluoride-induced chronic pancreatitis in rats: Role of anti-inflammatory, antioxidant, antifibrotic and anti-apoptotic activities. Int. Immunopharmacol. 2021, 93, 107372. [Google Scholar] [CrossRef]
- Molina-Jiménez, M.F.; Sánchez-Reus, M.I.; Andres, D.; Cascales, M.; Benedi, J. Neuroprotective effect of fraxetin and myricetin against rotenone-induced apoptosis in neuroblastoma cells. Brain Res. 2004, 1009, 9–16. [Google Scholar] [CrossRef]
- Sánchez-Reus, M.I.; Peinado, I.I.; Molina-Jiménez, M.F.; Benedí, J. Fraxetin prevents rotenone-induced apoptosis by induction of endogenous glutathione in human neuroblastoma cells. Neurosci. Res. 2005, 53, 48–56. [Google Scholar] [CrossRef]
- Li, Y.; Li, J.; Li, Y.; Wang, X.X.; Cao, A.C. Antimicrobial Constituents of the Leaves of Mikania micrantha H. B. K. PLoS ONE 2013, 8, e76725. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhao, Y.; Shi, Z.; Bai, Y. Antitumor effects of helenalin in doxorubicin-resistant leukemia cells are mediated via mitochondrial mediated apoptosis, loss of mitochondrial membrane potential, inhibition of cell migration and invasion and downregulation of PI3-kinase/AKT/m-TOR signa. J. Buon 2019, 24, 2068–2074. [Google Scholar] [PubMed]
- Moyo, P.; Kunyane, P.; Selepe, M.A.; Eloff, J.N.; Niemand, J.; Louw, A.I.; Maharaj, V.J.; Birkholtz, L.M. Bioassay-guided isolation and identification of gametocytocidal compounds from Artemisia afra (Asteraceae). Malar. J. 2019, 18, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramos, A.; Rivero, R.; Visozo, A.; Piloto, J.; García, A. Parthenin, a sesquiterpene lactone of Parthenium hysterophorus L. is a high toxicity clastogen. Mutat. Res.-Genet. Toxicol. Environ. Mutagen. 2002, 514, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S.; Matsuo, K.; Yaji, K.; Okajima-Miyazaki, S.; Harada, M.; Miyoshi, H.; Okamoto, Y.; Amamoto, T.; Shindo, M.; Omiecinski, C.J.; et al. (-)-Xanthatin selectively induces GADD45γ and stimulates caspase-independent cell death in human breast cancer MDA-MB-231 cells. Chem. Res. Toxicol. 2011, 24, 855–865. [Google Scholar] [CrossRef]
- Wu, Q.; Qin, Z.; Kuca, K.; You, L.; Zhao, Y.; Liu, A.; Musilek, K.; Chrienova, Z.; Nepovimova, E.; Oleksak, P.; et al. An update on T-2 toxin and its modified forms: Metabolism, immunotoxicity mechanism, and human exposure assessment. Arch. Toxicol. 2020, 94, 3645–3669. [Google Scholar] [CrossRef]
- Pai, J.T.; Hsu, C.Y.; Hua, K.T.; Yu, S.Y.; Huang, C.Y.; Chen, C.N.; Liao, C.H.; Weng, M.S. NBM-T-BBX-OS01, semisynthesized from osthole, induced G1 growth arrest through HDAC6 inhibition in lung cancer cells. Molecules 2015, 20, 8000–8019. [Google Scholar] [CrossRef] [Green Version]
- Tang, D.Z.; Hou, W.; Zhou, Q.; Zhang, M.; Holz, J.; Sheu, T.J.; Li, T.F.; Cheng, S.D.; Shi, Q.; Harris, S.E.; et al. Osthole stimulates osteoblast differentiation and bone formation by activation of β-catenin-BMP signaling. J. Bone Miner. Res. 2010, 25, 1234–1245. [Google Scholar] [CrossRef] [Green Version]
- You, L.; Feng, S.; An, R.; Wang, X. Osthole: A Promising Lead Compound for Drug Discovery from a Traditional Chinese Medicine (TCM). Nat. Prod. Commun. 2009, 4, 1934578X0900400. [Google Scholar] [CrossRef] [Green Version]
- Balaji, N.V.; Ramani, M.V.; Viana, A.G.; Sanglard, L.P.; White, J.; Mulabagal, V.; Lee, C.; Gana, T.J.; Egiebor, N.O.; Subbaraju, G.V.; et al. Design, synthesis and in vitro cell-based evaluation of the anti-cancer activities of hispolon analogs. Bioorganic Med. Chem. 2015, 23, 2148–2158. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.H.; Park, W.H. Pyrogallol-induced calf pulmonary arterial endothelial cell death via caspase-dependent apoptosis and GSH depletion. Food Chem. Toxicol. 2010, 48, 558–563. [Google Scholar] [CrossRef] [PubMed]
- Suh, D.Y.; Han, Y.N.; Han, B.H. Maltol, an antioxidant component of Korean red ginseng, shows little prooxidant activity. Arch. Pharm. Res. 1996, 19, 112–115. [Google Scholar] [CrossRef]
- Chemical Entities of Biological Interest Resveratrol CHEBI:27881. Available online: https://www.ebi.ac.uk/chebi/searchId.do?chebiId=CHEBI:27881 (accessed on 20 August 2022).
- Yang, C.J.; Wang, C.S.; Hung, J.Y.; Huang, H.W.; Chia, Y.C.; Wang, P.H.; Weng, C.F.; Huang, M.S. Pyrogallol induces G2-M arrest in human lung cancer cells and inhibits tumor growth in an animal model. Lung Cancer 2009, 66, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, V.; Darokar, M.P.; Fatima, A.; Kumar, J.K.; Chowdhury, C.; Saxena, H.O.; Dwivedi, G.R.; Shrivastava, K.; Gupta, V.; Chattopadhyay, S.K.; et al. Synthesis of diverse analogues of Oenostacin and their antibacterial activities. Bioorganic Med. Chem. 2007, 15, 518–525. [Google Scholar] [CrossRef] [PubMed]
- Andrioli, W.J.; Conti, R.; Araújo, M.J.; Zanasi, R.; Cavalcanti, B.C.; Manfrim, V.; Toledo, J.S.; Tedesco, D.; De Moraes, M.O.; Pessoa, C.; et al. Mycoleptones A-C and polyketides from the endophyte mycoleptodiscus indicus. J. Nat. Prod. 2014, 77, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Joo, S.H.; Lee, S.C.; Kim, S.K. UV absorbent, marmesin, from the bark of Thanakha, Hesperethusa crenulata L. J. Plant Biol. 2004, 47, 163–165. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. PubChem Compound Summary for CID 5282965, 9,10,13-Trihydroxy-11-octadecenoic Acid. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/5282965 (accessed on 20 August 2022).
- National Center for Biotechnology Information. PubChem Bioassay Record for AID 2300, Source: The Scripps Research Institute Molecular Screening Center. Available online: https://pubchem.ncbi.nlm.nih.gov/bioassay/2300 (accessed on 20 August 2022).
- National Center for Biotechnology Information. PubChem Compound Summary for CID 1052, Pyridoxamine. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Pyridoxamine (accessed on 20 August 2022).
- Kinoshita, G.; Nakamura, F.; Furuhata, Y. Inhibitory effects of Saposhnikovia root on CNS functions and peptic ulcers. J. Med. Pharm. Soc. Wakan-Yaku 1987, 4, 130–137. [Google Scholar]
- Chen, N.; Wu, Q.; Chi, G.; Soromou, L.W.; Hou, J.; Deng, Y.; Feng, H. Prime-O-glucosylcimifugin attenuates lipopolysaccharide-induced acute lung injury in mice. Int. Immunopharmacol. 2013, 16, 139–147. [Google Scholar] [CrossRef]
- Chang, C.Z.; Wu, S.C.; Kwan, A.L.; Lin, C.L. 4’-O-β-D-glucosyl-5-O-methylvisamminol, an active ingredient of Saposhnikovia divaricata, attenuates high-mobility group box 1 and subarachnoid hemorrhage-induced vasospasm in a rat model. Behav. Brain Funct. 2015, 11, 1–13. [Google Scholar] [CrossRef] [Green Version]
- National Center for Biotechnology Information. PubChem Compound Summary for CID 76318697, Yadanzioside D. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Yadanzioside-D (accessed on 24 August 2022).
- National Center for Biotechnology Information. PubChem Bioassay Record for AID 2330, Source: Broad Institute. Available online: https://pubchem.ncbi.nlm.nih.gov/bioassay/2330 (accessed on 21 August 2022).
- National Center for Biotechnology Information. PubChem Compound Summary for CID 5283137, Thromboxane B2. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/5283137 (accessed on 21 August 2022).
- National Center for Biotechnology Information. PubChem Compound Summary for CID 5280888, 6-Keto-Prostaglandin F1alpha. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/5280888 (accessed on 21 August 2022).
- Chen, B.H.; Wu, P.Y.; Chen, K.M.; Fu, T.F.; Wang, H.M.; Chen, C.Y. Antiallergic potential on RBL-2H3 cells of some phenolic constituents of Zingiber officinale (ginger). J. Nat. Prod. 2009, 72, 950–953. [Google Scholar] [CrossRef]
- Aun, L.L.; Azmi, M.N.; Ibrahim, H.; Awang, K.; Nagoor, N.H. 1′S-1′-acetoxyeugenol acetate: A novel phenylpropanoid from Alpinia conchigera enhances the apoptotic effects of paclitaxel in MCF-7 cells through NF-κB inactivation. Anticancer Drugs 2011, 22, 424–434. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information. PubChem Compound Summary for CID 28619, Precocene I. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Precocene-I (accessed on 24 August 2022).
- Musthafa, K.S.; Voravuthikunchai, S.P. Anti-virulence potential of eugenyl acetate against pathogenic bacteria of medical importance. Antonie van Leeuwenhoek. Int. J. Gen. Mol. Microbiol. 2015, 107, 703–710. [Google Scholar] [CrossRef] [PubMed]
- Kuca, K.; Dohnal, V.; Jezkova, A.; Jun, D. Metabolic Pathways of T-2 Toxin. Curr. Drug Metab. 2008, 9, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.R. Sesquiterpene lactones from Tithonia diversifolia act as peroxisome proliferator-activated receptor agonists. Bioorganic Med. Chem. Lett. 2012, 22, 2954–2958. [Google Scholar] [CrossRef]
- Emond, J.P.; Lacombe, L.; Caron, P.; Turcotte, V.; Simonyan, D.; Aprikian, A.; Saad, F.; Carmel, M.; Chevalier, S.; Guillemette, C.; et al. Urinary oestrogen steroidome as an indicator of the risk of localised prostate cancer progression. Br. J. Cancer 2021, 125, 78–84. [Google Scholar] [CrossRef]
- Li, S.; Chen, Y.; Xie, L.; Meng, Y.; Zhu, L.; Chu, H.; Gu, D.; Zhang, Z.; Du, M.; Wang, M. Sex hormones and genetic variants in hormone metabolic pathways associated with the risk of colorectal cancer. Environ. Int. 2020, 137, 105543. [Google Scholar] [CrossRef]
- Zhao, F.; Hao, Z.; Zhong, Y.; Xu, Y.; Guo, M.; Zhang, B.; Yin, X.; Li, Y.; Zhou, X. Discovery of breast cancer risk genes and establishment of a prediction model based on estrogen metabolism regulation. BMC Cancer 2021, 21, 1–11. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. PubChem Compound Summary for CID 23205, Helenalin. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Helenalin (accessed on 24 August 2022).
- Huang, D.-N.; Wang, S.; Sooranna, S.R.; Miao, J.-H. The Efficacy of Natural Bioactive Compounds for the Treatment of Nasopharyngeal Carcinoma. Mini-Rev. Med. Chem. 2021, 21, 1679–1691. [Google Scholar] [CrossRef]
- Picman, J.; Picman, A.K. Treatment of dermatitis from parthenin. Contact Dermat. 1985, 13, 9–13. [Google Scholar] [CrossRef]
- Huang, W.J.; Chen, C.C.; Chao, S.W.; Lee, S.S.; Hsu, F.L.; Lu, Y.L.; Hung, M.F.; Chang, C.I. Synthesis of n-hydroxycinnamides capped with a naturally occurring moiety as inhibitors of histone deacetylase. ChemMedChem 2010, 5, 598–607. [Google Scholar] [CrossRef]
- Zhang, C.; Ning, D.; Pan, J.; Chen, C.; Gao, C.; Ding, Z.; Jiang, F.; Li, M. Anti-Inflammatory Effect Fraction of Bletilla striata and Its Protective Effect on LPS-Induced Acute Lung Injury. Mediat. Inflamm. 2021, 2021, 6684120. [Google Scholar] [CrossRef] [PubMed]
- Pinkhien, T.; Petpiroon, N.; Sritularak, B.; Chanvorachote, P. Batatasin III inhibits migration of human lung cancer cells by suppressing epithelial to mesenchymal transition and FAK-AKT signals. Anticancer Res. 2017, 37, 6281–6289. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Nitya, A. Natural products. In Pharmacochemistry Library; Elsevier: Amsterdam, The Netherlands, 1997; Volume 25, pp. 71–123. [Google Scholar]
- Gaich, T.; Mulzer, J. 2.7 Chiral Pool Synthesis: Starting from Terpenes. In Comprehensive Chirality; Elsevier: Amsterdam, The Netherlands, 2012; Volume 2, pp. 163–206. ISBN 9780080951683. [Google Scholar]
- Dhakal, D.; Han, J.M.; Mishra, R.; Pandey, R.P.; Kim, T.S.; Rayamajhi, V.; Jung, H.J.; Yamaguchi, T.; Sohng, J.K. Characterization of Tailoring Steps of Nargenicin A1 Biosynthesis Reveals a Novel Analogue with Anticancer Activities. ACS Chem. Biol. 2020, 15, 1370–1380. [Google Scholar] [CrossRef]
- Coppock, R.W.; Christian, R.G.; Jacobsen, B.J. Aflatoxins. In Veterinary Toxicology: Basic and Clinical Principles, 3rd ed.; Academic Press: Cambridge, MA, USA, 2018; pp. 983–994. ISBN 9780128114100. [Google Scholar]
- Agahi, F.; Juan-García, A.; Font, G.; Juan, C. Study of enzymatic activity in human neuroblastoma cells SH-SY5Y exposed to zearalenone’s derivates and beauvericin. Food Chem. Toxicol. 2021, 152, 112227. [Google Scholar] [CrossRef] [PubMed]
- MeSH Pharmacological Classification Antimetabolites, Antineoplastic-MeSH-NCBI. Available online: https://www.ncbi.nlm.nih.gov/mesh/68000964 (accessed on 27 October 2022).
- National Center for Biotechnology Information. PubChem Compound Summary for CID 3840, Kojic Acid. Retrieved 24 August 2022. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Kojic-acid (accessed on 27 October 2022).
- Chemical Entities of Biological Interest Ferulic acid CHEBI:17620. Retrieved 20 August 2022. Available online: https://www.ebi.ac.uk/chebi/searchId.do?chebiId=CHEBI:17620 (accessed on 27 October 2022).
- Chemical Entities of Biological Interest Scoparone CHEBI:9055. Retrieved 20 August 2022. Available online: https://www.ebi.ac.uk/chebi/searchId.do?chebiId=CHEBI:9055 (accessed on 27 October 2022).
- Chemical Entities of Biological Interest Fraxetin CHEBI:5169. Retrieved 20 August 2022. Available online: https://www.ebi.ac.uk/chebi/searchId.do?chebiId=CHEBI:5169 (accessed on 27 October 2022).
- Michalak, I.; Chojnacka, K. Algae as production systems of bioactive compounds. Eng. Life Sci. 2015, 15, 160–176. [Google Scholar] [CrossRef]
- Leandro, A.; Pereira, L.; Gonçalves, A.M.M. Diverse Applications of Marine Macroalgae. Mar. Drugs 2019, 18, 17. [Google Scholar] [CrossRef]
- Zhang, G.; Li, J.; Zhu, T.; Gu, Q.; Li, D. Advanced tools in marine natural drug discovery. Curr. Opin. Biotechnol. 2016, 42, 13–23. [Google Scholar] [CrossRef]
- Suleria, H.A.R.; Gobe, G.; Masci, P.; Osborne, S.A. Marine bioactive compounds and health promoting perspectives; innovation pathways for drug discovery. Trends Food Sci. Technol. 2016, 50, 44–55. [Google Scholar] [CrossRef]
- Sigwart, J.D.; Blasiak, R.; Jaspars, M.; Jouffray, J.-B.; Tasdemir, D. Unlocking the potential of marine biodiscovery. Nat. Prod. Rep. 2021, 38, 1235–1242. [Google Scholar] [CrossRef]
- Petras, D.; Koester, I.; Da Silva, R.; Stephens, B.M.; Haas, A.F.; Nelson, C.E.; Kelly, L.W.; Aluwihare, L.I.; Dorrestein, P.C. High-resolution liquid chromatography tandem mass spectrometry enables large scale molecular characterization of dissolved organic matter. Front. Mar. Sci. 2017, 4, 405. [Google Scholar] [CrossRef] [Green Version]
- Sogin, E.M.; Putnam, H.M.; Nelson, C.E.; Anderson, P.; Gates, R.D. Correspondence of coral holobiont metabolome with symbiotic bacteria, archaea and Symbiodinium communities. Environ. Microbiol. Rep. 2017, 9, 310–315. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, E.M.; Sobus, J.R.; Grulke, C.M.; Richard, A.M.; Newton, S.R.; Strynar, M.J.; Mansouri, K.; Williams, A.J. EPA’s non-targeted analysis collaborative trial (ENTACT): Genesis, design, and initial findings. Anal. Bioanal. Chem. 2019, 411, 853–866. [Google Scholar] [CrossRef] [PubMed]
- Hollender, J.; Schymanski, E.L.; Singer, H.P.; Ferguson, P.L. Nontarget Screening with High Resolution Mass Spectrometry in the Environment: Ready to Go? Environ. Sci. Technol. 2017, 51, 11505–11512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marshall, A.G.; Hendrickson, C.L.; Jackson, G.S. Fourier transform ion cyclotron resonance mass spectrometry: A primer. Mass Spectrom. Rev. 1998, 17, 1–35. [Google Scholar] [CrossRef]
- Nikolaev, E.N.; Kostyukevich, Y.I.; Vladimirov, G.N. Fourier transform ion cyclotron resonance (FT ICR) mass spectrometry: Theory and simulations. Mass Spectrom. Rev. 2016, 35, 219–258. [Google Scholar] [CrossRef]
- Zark, M.; Christoffers, J.; Dittmar, T. Molecular properties of deep-sea dissolved organic matter are predictable by the central limit theorem: Evidence from tandem FT-ICR-MS. Mar. Chem. 2017, 191, 9–15. [Google Scholar] [CrossRef]
- Hawkes, J.A.; Patriarca, C.; Sjöberg, P.J.R.; Tranvik, L.J.; Bergquist, J. Extreme isomeric complexity of dissolved organic matter found across aquatic environments. Limnol. Oceanogr. Lett. 2018, 3, 21–30. [Google Scholar] [CrossRef] [Green Version]
- Regal, A.L.; Alves, V.; Gomes, R.; Matos, J.; Bandarra, N.M.; Afonso, C.; Cardoso, C. Drying process, storage conditions, and time alter the biochemical composition and bioactivity of the anti-greenhouse seaweed Asparagopsis taxiformis. Eur. Food Res. Technol. 2020, 246, 781–793. [Google Scholar] [CrossRef]
- Nunes, N.; Ferraz, S.; Valente, S.; Barreto, M.C.; Pinheiro de Carvalho, M.A.A. Biochemical composition, nutritional value, and antioxidant properties of seven seaweed species from the Madeira Archipelago. J. Appl. Phycol. 2017, 29, 2427–2437. [Google Scholar] [CrossRef]
- Jard, G.; Marfaing, H.; Carrère, H.; Delgenes, J.P.; Steyer, J.P.; Dumas, C. French Brittany macroalgae screening: Composition and methane potential for potential alternative sources of energy and products. Bioresour. Technol. 2013, 144, 492–498. [Google Scholar] [CrossRef]
- Bogaert, K.A.; Delva, S.; De Clerck, O. Concise review of the genus Dictyota J.V. Lamouroux. J. Appl. Phycol. 2020, 32, 1521–1543. [Google Scholar] [CrossRef]
- Zubia, M.; Robledo, D.; Freile-Pelegrin, Y. Antioxidant activities in tropical marine macroalgae from the Yucatan Peninsula, Mexico. J. Appl. Phycol. 2007, 19, 449–458. [Google Scholar] [CrossRef]
- Vega, J.; Álvarez-Gómez, F.; Güenaga, L.; Figueroa, F.L.; Gómez-Pinchetti, J.L. Antioxidant activity of extracts from marine macroalgae, wild-collected and cultivated, in an integrated multi-trophic aquaculture system. Aquaculture 2020, 522, 735088. [Google Scholar] [CrossRef]
- Connan, S.; Goulard, F.; Stiger, V.; Deslandes, E.; Gall, E.A. Interspecific and temporal variation in phlorotannin levels in an assemblage of brown algae. Bot. Mar. 2004, 47, 410–416. [Google Scholar] [CrossRef]
- Celis-Plá, P.S.M.; Korbee, N.; Gómez-Garreta, A.; Figueroa, F.L. Seasonal photoacclimation patterns in the intertidal macroalga Cystoseira tamariscifolia (Ochrophyta). Sci. Mar. 2014, 78, 377–388. [Google Scholar] [CrossRef] [Green Version]
- Celis-Plá, P.S.M.; Brown, M.T.; Santillán-Sarmiento, A.; Korbee, N.; Sáez, C.A.; Figueroa, F.L. Ecophysiological and metabolic responses to interactive exposure to nutrients and copper excess in the brown macroalga Cystoseira tamariscifolia. Mar. Pollut. Bull. 2018, 128, 214–222. [Google Scholar] [CrossRef]
- Schneider, G.; Figueroa, F.L.; Vega, J.; Chaves, P.; Álvarez-Gómez, F.; Korbee, N.; Bonomi-Barufi, J. Photoprotection properties of marine photosynthetic organisms grown in high ultraviolet exposure areas: Cosmeceutical applications. Algal Res. 2020, 49, 101956. [Google Scholar] [CrossRef]
- Bridi, R.; Troncoso, M.J.; Folch-Cano, C.; Fuentes, J.; Speisky, H.; López-Alarcón, C. A Polyvinylpolypyrrolidone (PVPP)-Assisted Folin–Ciocalteu Assay to Assess Total Phenol Content of Commercial Beverages. Food Anal. Methods 2014, 7, 2075–2083. [Google Scholar] [CrossRef]
- Parys, S.; Rosenbaum, A.; Kehraus, S.; Reher, G.; Glombitza, K.-W.; König, G.M. Evaluation of Quantitative Methods for the Determination of Polyphenols in Algal Extracts. J. Nat. Prod. 2007, 70, 1865–1870. [Google Scholar] [CrossRef]
- Kumar, M.; Kuzhiumparambil, U.; Pernice, M.; Jiang, Z.; Ralph, P.J. Metabolomics: An emerging frontier of systems biology in marine macrophytes. Algal Res. 2016, 16, 76–92. [Google Scholar] [CrossRef]
- De Paula, J.C.; Cavalcanti, D.N.; Yoneshigue-Valentin, Y.; Teixeira, V.L. Diterpenes from marine brown alga Dictyota guineensis (Dictyotaceae, Phaeophyceae). Rev. Bras. Farmacogn. 2012, 22, 736–740. [Google Scholar] [CrossRef] [Green Version]
- Hamid, S.S.; Wakayama, M.; Ichihara, K.; Sakurai, K.; Ashino, Y.; Kadowaki, R.; Soga, T.; Tomita, M. Metabolome profiling of various seaweed species discriminates between brown, red, and green algae. Planta 2019, 249, 1921–1947. [Google Scholar] [CrossRef]
- Gaubert, J.; Greff, S.; Thomas, O.P.; Payri, C.E. Metabolomic variability of four macroalgal species of the genus Lobophora using diverse approaches. Phytochemistry 2019, 162, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Nylund, G.M.; Weinberger, F.; Rempt, M.; Pohnert, G. Metabolomic assessment of induced and activated chemical defence in the invasive red alga gracilaria vermiculophylla. PLoS ONE 2011, 6, e29359. [Google Scholar] [CrossRef] [PubMed]
- Dixit, D.; Reddy, C.R.K.; Trivedi, M.H.; Gadhavi, D.K. Non-targeted metabolomics approach to assess the brown marine macroalga Dictyota dichotoma as a functional food using liquid chromatography with mass spectrometry. Sep. Sci. Plus 2020, 3, 140–149. [Google Scholar] [CrossRef]
- Mateos, R.; Pérez-Correa, J.; Domínguez, H. Bioactive properties of marine phenolics. Mar. Drugs 2020, 18, 501. [Google Scholar] [CrossRef]
- Santos, S.A.O.; Félix, R.; Pais, A.C.S.; Rocha, S.M.; Silvestre, A.J.D. The Quest for Phenolic Compounds from Macroalgae: A Review of Extraction and Identification Methodologies. Biomolecules 2019, 9, 847. [Google Scholar] [CrossRef] [Green Version]
- Pinto, D.C.G.A.; Lesenfants, M.L.; Rosa, G.P.; Barreto, M.C.; Silva, A.M.S.; Seca, A.M.L. GC-and UHPLC-MS Profiles as a Tool to Valorize the Red Alga Asparagopsis armata. Appl. Sci. 2022, 12, 892. [Google Scholar] [CrossRef]
- Zhong, B.; Robinson, N.A.; Warner, R.D.; Barrow, C.J.; Dunshea, F.R.; Suleria, H.A.R. LC-ESI-QTOF-MS/MS Characterization of Seaweed Phenolics and Their Antioxidant Potential. Mar. Drugs 2020, 18, 331. [Google Scholar] [CrossRef]
- Chauhan, V.; Chauhan, A. Oxidative stress in Alzheimer’s disease. Pathophysiology 2006, 13, 195–208. [Google Scholar] [CrossRef]
- Thanan, R.; Oikawa, S.; Hiraku, Y.; Ohnishi, S.; Ma, N.; Pinlaor, S.; Yongvanit, P.; Kawanishi, S.; Murata, M.; Thanan, R.; et al. Oxidative Stress and Its Significant Roles in Neurodegenerative Diseases and Cancer. Int. J. Mol. Sci. 2014, 16, 193–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blesa, J.; Trigo-Damas, I.; Quiroga-Varela, A.; Jackson-Lewis, V.R. Oxidative stress and Parkinson’s disease. Front. Neuroanat. 2015, 9, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siti, H.N.; Kamisah, Y.; Kamsiah, J. The role of oxidative stress, antioxidants and vascular inflammation in cardiovascular disease (a review). Vascul. Pharmacol. 2015, 71, 40–56. [Google Scholar] [CrossRef] [PubMed]
- Félix, R.; Dias, P.; Félix, C.; Cerqueira, T.; Andrade, P.B.; Valentão, P.; Lemos, M.F.L. The biotechnological potential of Asparagopsis armata: What is known of its chemical composition, bioactivities and current market? Algal Res. 2021, 60, 102534. [Google Scholar] [CrossRef]
- Mekinić, I.G.; Šimat, V.; Botić, V.; Crnjac, A.; Smoljo, M.; Soldo, B.; Ljubenkov, I.; Čagalj, M.; Skroza, D. Bioactive phenolic metabolites from adriatic brown algae Dictyota dichotoma and padina pavonica (Dictyotaceae). Foods 2021, 10, 1187. [Google Scholar] [CrossRef]
- Martins, C.D.L.; Ramlov, F.; Nocchi Carneiro, N.P.; Gestinari, L.M.; dos Santos, B.F.; Bento, L.M.; Lhullier, C.; Gouvea, L.; Bastos, E.; Horta, P.A.; et al. Antioxidant properties and total phenolic contents of some tropical seaweeds of the Brazilian coast. J. Appl. Phycol. 2013, 25, 1179–1187. [Google Scholar] [CrossRef]
- El-Shenody, R.A.; Ashour, M.; Ghobara, M.M.E. Evaluating the chemical composition and antioxidant activity of three Egyptian seaweeds: Dictyota dichotoma, Turbinaria decurrens, and Laurencia obtusa. Braz. J. Food Technol. 2019, 22. [Google Scholar] [CrossRef] [Green Version]
- Platsidaki, E.; Dessinioti, C. Recent advances in understanding Propionibacterium acnes (Cutibacterium acnes) in acne. F1000Research 2018, 7, 1953. [Google Scholar] [CrossRef] [Green Version]
- Shanmughapriya, S.; Manilal, A.; Sujith, S.; Selvin, J.; Kiran, G.S.; Natarajaseenivasan, K. Antimicrobial activity of seaweeds extracts against multiresistant pathogens. Ann. Microbiol. 2008, 58, 535–541. [Google Scholar] [CrossRef]
- Tuney, I.; Cadirci, B.H.; Unal, D.; Sukatar, A. Locational and organic solvent variation in antimicrobial activities of crude extracts of marine algae from the coast of Izmir (Turkey). Fresenius Environ. Bull. 2007, 16, 428–434. [Google Scholar]
- Lee, M.S.; Yim, M.-J.; Lee, J.M.; Lee, D.-S.; Kim, M.-Y.; Eom, S.-H. In vitro Antimicrobial Activities of Edible Seaweeds Extracts Against Cutibacterium acnes. Korean J. Fish. Aquat. Sci. 2021, 54, 111–117. [Google Scholar] [CrossRef]
- Pineau, R.M.; Hanson, S.E.; Lyles, J.T.; Quave, C.L. Growth inhibitory activity of callicarpa americana leaf extracts against Cutibacterium acnes. Front. Pharmacol. 2019, 10, 1206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Canha, M.N.; Komarnytsky, S.; Langhansova, L.; Lall, N. Exploring the Anti-Acne Potential of Impepho [Helichrysum odoratissimum (L.) Sweet] to Combat Cutibacterium acnes Virulence. Front. Pharmacol. 2020, 10, 1559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ina-Salwany, M.Y.; Al-saari, N.; Mohamad, A.; Mursidi, F.A.; Mohd-Aris, A.; Amal, M.N.A.; Kasai, H.; Mino, S.; Sawabe, T.; Zamri-Saad, M. Vibriosis in Fish: A Review on Disease Development and Prevention. J. Aquat. Anim. Health 2019, 31, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Álvarez, C.; Santos, Y. Identification and typing of fish pathogenic species of the genus Tenacibaculum. Appl. Microbiol. Biotechnol. 2018, 102, 9973–9989. [Google Scholar] [CrossRef]
- Shannon, E.; Abu-Ghannam, N. Antibacterial Derivatives of Marine Algae: An Overview of Pharmacological Mechanisms and Applications. Mar. Drugs 2016, 14, 81. [Google Scholar] [CrossRef]
- Cardoso, S.L.; Costa, C.S.D.; Nishikawa, E.; da Silva, M.G.C.; Vieira, M.G.A. Biosorption of toxic metals using the alginate extraction residue from the brown algae Sargassum filipendula as a natural ion-exchanger. J. Clean. Prod. 2017, 165, 491–499. [Google Scholar] [CrossRef]
- Varaprasad, K.; Jayaramudu, T.; Kanikireddy, V.; Toro, C.; Sadiku, E.R. Alginate-based composite materials for wound dressing application:A mini review. Carbohydr. Polym. 2020, 236, 116025. [Google Scholar] [CrossRef]
- Wang, Y.; Xing, M.; Cao, Q.; Ji, A.; Liang, H.; Song, S. Biological Activities of Fucoidan and the Factors Mediating Its Therapeutic Effects: A Review of Recent Studies. Mar. Drugs 2019, 17, 183. [Google Scholar] [CrossRef] [Green Version]
- Jönsson, M.; Allahgholi, L.; Sardari, R.R.R.; Hreggvidsson, G.O.; Nordberg Karlsson, E. Extraction and Modification of Macroalgal Polysaccharides for Current and Next-Generation Applications. Molecules 2020, 25, 930. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.W.; Lee, J.; Park, S.I.; Shin, M.S. Antimicrobial, Antioxidative, Elastase and Tyrosinase Inhibitory Effect of Supercritical and Hydrothermal Halopteris scoparia Extract. Turk. J. Comput. Math. Educ. 2021, 12, 407–413. [Google Scholar] [CrossRef]
- Mercado, J.M.; Gómez-Jakobsen, F.; Korbee, N.; Aviles, A.; Bonomi-Barufi, J.; Muñoz, M.; Reul, A.; Figueroa, F.L. Analyzing environmental factors that favor the growth of the invasive brown macroalga Rugulopteryx okamurae (Ochrophyta): The probable role of the nutrient excess. Mar. Pollut. Bull. 2022, 174, 113315. [Google Scholar] [CrossRef] [PubMed]
- Lourenço, S.O.; Barbarino, E.; De-Paula, J.C.; Pereira, L.O.D.S.; Lanfer Marquez, U.M. Amino acid composition, protein content and calculation of nitrogen-to-protein conversion factors for 19 tropical seaweeds. Phycol. Res. 2002, 50, 233–241. [Google Scholar] [CrossRef]
- DuBois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric Method for Determination of Sugars and Related Substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Folch, J.; Lees, M.; Sloan Stanley, G. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]
- Singleton, V.; Rossi, J. Colorimetry of total phenolics with phosphomolybdic-phosphotungstic acid reagents. Am. J. Enol. Vitic. 1965, 16, 144–158. [Google Scholar]
- Van Alstyne, K.L. Comparison of three methods for quantifying brown algal polyphenolic compounds. J. Chem. Ecol. 1995, 21, 45–58. [Google Scholar] [CrossRef]
- Hansman, R.L.; Dittmar, T.; Herndl, G.J. Conservation of dissolved organic matter molecular composition during mixing of the deep water masses of the northeast Atlantic Ocean. Mar. Chem. 2015, 177, 288–297. [Google Scholar] [CrossRef]
- Osterholz, H.; Singer, G.; Wemheuer, B.; Daniel, R.; Simon, M.; Niggemann, J.; Dittmar, T. Deciphering associations between dissolved organic molecules and bacterial communities in a pelagic marine system. ISME J. 2016, 10, 1717–1730. [Google Scholar] [CrossRef] [Green Version]
- Merder, J.; Freund, J.A.; Feudel, U.; Hansen, C.T.; Hawkes, J.A.; Jacob, B.; Klaproth, K.; Niggemann, J.; Noriega-Ortega, B.E.; Osterholz, H.; et al. ICBM-OCEAN: Processing Ultrahigh-Resolution Mass Spectrometry Data of Complex Molecular Mixtures. Anal. Chem. 2020, 92, 6832–6838. [Google Scholar] [CrossRef]
- Merder, J.; Freund, J.A.; Feudel, U.; Niggemann, J.; Singer, G.; Dittmar, T. Improved mass accuracy and isotope confirmation through alignment of ultrahigh-resolution mass spectra of complex natural mixtures. Anal. Chem. 2020, 92, 2558–2565. [Google Scholar] [CrossRef] [PubMed]
- Koch, B.P.; Dittmar, T. From mass to structure: An aromaticity index for high-resolution mass data of natural organic matter. Rapid Commun. Mass Spectrom. 2006, 20, 926–932. [Google Scholar] [CrossRef] [Green Version]
- The Pallets Projects Welcome to Flask—Flask Documentation (2.0.x). 2010. Available online: https://flask.palletsprojects.com/en/2.0.x/ (accessed on 2 November 2022).
- Reitz, K.; Benfield, C.; Cordasco, I.; Prewitt, N.; Larson, S.M. (2022) Requests: HTTP for humans. A Kenneth Reitz Project. 2021. Available online: https://docs.python-requests.org/en/master/ (accessed on 2 November 2022).
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef] [PubMed]
- Haug, K.; Cochrane, K.; Nainala, V.C.; Williams, M.; Chang, J.; Jayaseelan, K.V.; O’Donovan, C. MetaboLights: A resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 2020, 48, D440–D444. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Thiessen, P.A.; Cheng, T.; Yu, B.; Bolton, E.E. An update on PUG-REST: RESTful interface for programmatic access to PubChem. Nucleic Acids Res. 2018, 46, W563–W570. [Google Scholar] [CrossRef]
- European Bioinformatics Institute MetaboLights RESTful WebService. Available online: https://www.ebi.ac.uk/metabolights/ws/api/spec.html#!/spec (accessed on 2 November 2022).
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Brand-Williams, W.; Cuvelier, M.E.; Berset, C. Use of a free radical method to evaluate antioxidant activity. LWT-Food Sci. Technol. 1995, 28, 25–30. [Google Scholar] [CrossRef]
- García-Márquez, J.; Barany, A.; Ruiz, Á.B.; Costas, B.; Arijo, S.; Mancera, J.M. Antimicrobial and toxic activity of citronella essential oil (Cymbopogon nardus), and its effect on the growth and metabolism of gilthead seabream (sparus aurata l.). Fishes 2021, 6, 61. [Google Scholar] [CrossRef]
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; Al, E. Community Ecology Package [R package vegan version 2.6-4]. 2022. Available online: https://cran.r-project.org/web/packages/vegan/vegan.pdf (accessed on 2 November 2022).





| A. armata | R. okamurae | |
|---|---|---|
| Organic matter | 58.2 ± 1.4 | 72.2 ± 1.4 |
| Ashes | 41.8 ± 1.4 | 27.8 ± 1.4 |
| Carbon | 19.0 ± 0.9 | 35.3 ± 0.3 |
| Nitrogen | 2.3 ± 0.3 | 1.7 ± 0.1 |
| C:N | 8.6 ± 0.7 | 19.9 ± 1.0 |
| Sulphur | 1.9 ± 0.3 | 0.8 ± 0.1 |
| Total proteins | 10.56 ± 0.8 | 9.15 ± 0.6 |
| Carbohydrates | 13.73 ± 2.1 | 8.8 ± 0.4 |
| Lipids | 2.5 ± 0.1 | 8.0 ± 0.4 |
| R. okamurae | A. armata | |||||
|---|---|---|---|---|---|---|
| dH2O | dH2O:EtOH (1:4) | dH2O:MeOH (1:4) | dH2O | dH2O:EtOH (1:4) | dH2O:MeOH (1:4) | |
| HUMAN | ||||||
| S. aureus | - | 25 | 25 | - | 25 | 25 |
| E. coli | - | - | - | - | - | - |
| P. aeruginosa | - | - | - | - | 50 | 50 |
| S. enterica | - | - | - | - | - | - |
| E. faecium | - | - | - | - | - | - |
| C. acnes | - | 50 | 50 | - | - | - |
| FISH | ||||||
| V. anguillarum | - | - | - | 25 | 6.25 | 12.5 |
| V. harveyi | - | - | - | 50 | - | - |
| P. damselae subsp. piscicida | - | - | - | - | 50 | 50 |
| T. maritimum | - | - | - | 25 | - | - |
| T. soleae | - | - | - | 12.5 | 12.5 | 12.5 |
| T. gallaecicum | - | - | - | 12.5 | 12.5 | 12.5 |
| A. hydrophila | - | - | - | 25 | 25 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vega, J.; Catalá, T.S.; García-Márquez, J.; Speidel, L.G.; Arijo, S.; Cornelius Kunz, N.; Geisler, C.; Figueroa, F.L. Molecular Diversity and Biochemical Content in Two Invasive Alien Species: Looking for Chemical Similarities and Bioactivities. Mar. Drugs 2023, 21, 5. https://doi.org/10.3390/md21010005
Vega J, Catalá TS, García-Márquez J, Speidel LG, Arijo S, Cornelius Kunz N, Geisler C, Figueroa FL. Molecular Diversity and Biochemical Content in Two Invasive Alien Species: Looking for Chemical Similarities and Bioactivities. Marine Drugs. 2023; 21(1):5. https://doi.org/10.3390/md21010005
Chicago/Turabian StyleVega, Julia, Teresa S. Catalá, Jorge García-Márquez, Linn G. Speidel, Salvador Arijo, Niklas Cornelius Kunz, Christoph Geisler, and Félix L. Figueroa. 2023. "Molecular Diversity and Biochemical Content in Two Invasive Alien Species: Looking for Chemical Similarities and Bioactivities" Marine Drugs 21, no. 1: 5. https://doi.org/10.3390/md21010005