Monarubins A–C from the Marine Shellfish-Associated Fungus Monascus ruber BB5
Abstract
:1. Introduction
2. Results and Discussion
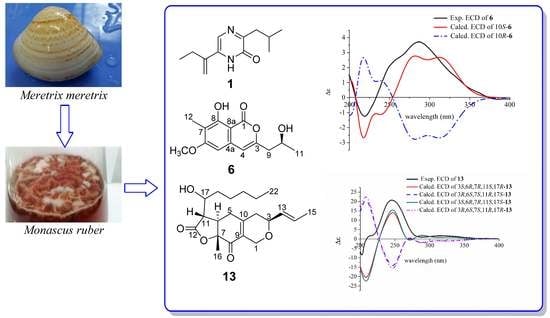
2.1. Structural Elucidation
2.2. Biological Activity
3. Materials and Methods
3.1. General Procedures
3.2. Fungal Strain and Culture Method
3.3. Extraction and Isolation
3.4. Computational Methods
3.5. X-ray Crystallographic Analysis for 4
3.6. Cytotoxicity Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zhao, W.W.; Shen, H.H. A statistical analysis of China’s fisheries in the 12th five-year period. Aquac. Fish. 2016, 1, 41–49. [Google Scholar] [CrossRef]
- Ferreira, J.G.; Sequeira, A.; Hawkins, A.J.S.; Newton, A.; Nickell, T.D.; Pastres, R.; Forte, J.; Bodoy, A.; Bricker, S.B. Analysis of coastal and offshore aquaculture: Application of the FARM model to multiple systems and shellfish species. Aquaculture 2009, 289, 32–41. [Google Scholar] [CrossRef]
- Shapiro, K.; Silver, M.; Byrne, B.A.; Berardi, T.; Aguilar, B.; Melli, A.; Smith, W.A. Fecal indicator bacteria and zoonotic pathogens in marine snow and California mussels (Mytilus californianus). FEMS Microbiol. Ecol. 2018, 94, 172. [Google Scholar] [CrossRef] [PubMed]
- Salam, M.A.; Paul, S.C.; Noor, S.N.B.M.; Siddiqua, S.A.; Aka, T.D.; Wahab, R.; Aweng, E.R. Contamination profile of heavy metals in marine fish and shellfish. Glob. J. Environ. Sci. Manag. 2019, 5, 225–236. [Google Scholar]
- Hallegaeff, G.M. A review of harmful algal blooms and their apparent global increase. Phycologia 1993, 2, 79–99. [Google Scholar] [CrossRef] [Green Version]
- Natarajan, S.B.; Kim, Y.S.; Hwang, J.W.; Park, P.J. Immunomodulatory properties of shellfish derivatives associated with human health. RSC Adv. 2016, 6, 26163–26177. [Google Scholar] [CrossRef]
- Ma, J.Y.; Li, Y.G.; Ye, Q.; Li, J.; Hua, Y.J.; Ju, D.J.; Zhang, D.C.; Cooper, R.; Chang, M. Constituents of red yeast rice, a traditional Chinese food and medicine. J. Agric. Food Chem. 2000, 48, 5220–5225. [Google Scholar] [CrossRef]
- Wu, H.C.; Cheng, M.J.; Wu, M.D.; Chen, J.J.; Chen, Y.L.; Chang, H.S. Three new constituents from the fungus of Monascus purpureus and their anti-inflammatory activity. Phytochem. Lett. 2019, 31, 242–248. [Google Scholar] [CrossRef]
- Akihisa, T.; Tokuda, H.; Yasukawa, K.; Ukiya, M.; Kiyota, A.; Sakamoto, N.; Suzuki, T.; Tanabe, N.; Nishino, H. Azaphilones, furanoisophthalides, and amino acids from the extracts of Monascus pilosus-fermented rice red-mold rice and their chemopreventive effects. J. Agric. Food Chem. 2005, 53, 562–565. [Google Scholar] [CrossRef]
- Wei, W.D.; Lin, S.; Chen, M.H.; Liu, T.X.; Wang, A.; Li, J.J.; Guo, Q.L.; Shang, X.Y. Monascustin, an unusual γ-lactam from red yeast rice. J. Nat. Prod. 2017, 80, 201–204. [Google Scholar] [CrossRef]
- Huang, Z.B.; Xu, Y.; Li, L.S.; Li, Y.P. Two new Monascus metabolites with strong blue fluorescence isolated from red yeast rice. J. Agric. Food Chem. 2008, 56, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Patakova, P. Monascus secondary metabolites: Production and biological activity. J. Ind. Microbiol. Biot. 2013, 40, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Hong, M.Y.; Seeram, N.P.; Zhang, Y.J.; Heber, D. Anticancer effects of Chinese red yeast rice versus monacolin K alone on colon cancer cells. J. Nutr. Biochem. 2008, 19, 448–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, M.J.; Wu, M.D.; Chen, I.S.; Tseng, M.; Yuan, G.F. Chemical constituents from the fungus Monascus purpureus and their antifungal activity. Phytochem. Lett. 2011, 4, 372–376. [Google Scholar] [CrossRef]
- Hsu, L.C.; Liang, Y.H.; Hsu, Y.W.; Kuo, Y.H.; Pan, T.M. Anti-inflammatory properties of yellow and orange pigments from Monascus purpureus NTU 568. J. Agric. Food Chem. 2013, 61, 2796–2802. [Google Scholar] [CrossRef]
- Lee, C.L.; Wang, J.J.; Pan, T.M. Red mold rice extract represses amyloid beta peptide-induced neurotoxicity via potent synergism of anti-inflammatory and antioxidative effect. Appl. Microbiol. Biotechnol. 2008, 79, 829–841. [Google Scholar] [CrossRef]
- Pandey, V.V.; Varshney, V.K.; Pandey, A. Lovastatin: A journey from ascomycetes to basidiomycetes fungi. J. Biol. Act. Product. Nat. 2019, 9, 162–178. [Google Scholar] [CrossRef]
- Li, H.J.; Lan, W.J.; Lam, C.K.; Yang, F.; Zhu, X.F. Hirsutane sesquiterpenoids from the marine-derived fungus Chondrostereum sp. Chem. Biodivers. 2011, 8, 317–324. [Google Scholar] [CrossRef]
- Li, H.J.; Chen, T.; Xie, Y.L.; Chen, W.D.; Zhu, X.F.; Lan, W.J. Isolation and structure elucidation of chondrosterins F–H from the marine fungus Chondrostereum sp. Mar. Drugs 2013, 11, 551–558. [Google Scholar] [CrossRef] [Green Version]
- Li, H.J.; Xie, Y.L.; Xie, Z.L.; Chen, Y.; Lam, C.K.; Lan, W.J. Chondrosterins A–E, triquinane-type sesquiterpenoids from soft coral-associated fungus Chondrostereum sp. Mar. Drugs 2012, 10, 627–638. [Google Scholar] [CrossRef] [Green Version]
- Li, H.J.; Jiang, W.H.; Liang, W.L.; Huang, J.X.; Mo, Y.F.; Ding, Y.Q.; Lam, C.K.; Qian, X.J.; Zhu, X.F.; Lan, W.J. Induced marine fungus Chondrostereum sp. as a means of producing new sesquiterpenoids chondrosterins I and J by using glycerol as the carbon source. Mar. Drugs 2014, 12, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Lan, W.J.; Deng, R.; Feng, G.K.; Xu, Q.Y.; Hu, Z.Y.; Zhu, X.F.; Li, H.J. Additional new cytotoxic triquinane-type sesquiterpenoids chondrosterins K–M from the marine fungus Chondrostereum sp. Mar. Drugs 2016, 14, 157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lan, W.J.; Liu, W.; Liang, W.L.; Xu, Z.; Le, X.; Xu, J.; Lam, C.K.; Yang, D.P.; Li, H.J.; Wang, L.Y. Pseudaboydins A and B, novel isobenzofuranone derivatives from marine fungus Pseudallescheria boydii associated with starfish Acanthaster planci. Mar. Drugs 2014, 12, 4188–4199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, L.H.; Xu, M.Y.; Li, H.J.; Li, J.Q.; Chen, Y.X.; Ma, W.Z.; Li, Y.P.; Xu, J.; Yang, D.P.; Lan, W.J. Amino acid-directed strategy for inducing the marine-derived fungus Scedosporium apiospermum F41–1 to maximize alkaloid diversity. Org. Lett. 2017, 19, 4888–4891. [Google Scholar] [CrossRef]
- Cao, Q.X.; Wei, J.H.; Deng, R.; Feng, G.K.; Zhu, X.F.; Lan, W.J.; Li, H.J. Two new pyripyropenes from the marine fungus Fusarium sp. 2016F18-1. Chem. Biodivers. 2017, 14, e1600298. [Google Scholar] [CrossRef]
- Yuan, M.X.; Qiu, Y.; Ran, Y.Q.; Feng, G.K.; Deng, R.; Zhu, X.F.; Lan, W.J.; Li, H.J. Exploration of indole alkaloids from marine fungus Pseudallescheria boydii F44-1 using an amino acid-directed strategy. Mar. Drugs 2019, 17, 77. [Google Scholar] [CrossRef] [Green Version]
- Dutcher, J.D. Aspergillic acid: An antibiotic substance produced by Aspergillus flavus. III. Structure of hydroxyaspergillic acid. J. Biol. Chem. 1958, 232, 785–795. [Google Scholar]
- Sasaki, M.; Asao, Y.; Yokotsuka, T. Compounds produced by molds. III. Fluorescent compounds produced by Japanese commercial molds. Nippon Nogeikagaku Kaishi 1968, 42, 288–293. [Google Scholar] [CrossRef] [Green Version]
- Li, H.J.; Cai, Y.T.; Chen, Y.Y.; Lam, C.K.; Lan, W.J. Metabolites of marine fungus Aspergillus sp. collected from soft coral Sarcophyton tortuosum. Chem. Res. Chin. Univ. 2010, 26, 415–419. [Google Scholar]
- Alvi, K.A.; Casey, A.; Nair, B.G. Pulchellalactam: A CD45 protein tyrosine phosphatase inhibitor from the marine fungus Corollospora pulchella. J. Antibiot. 1997, 51, 515–517. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, Y.; Masuma, R.; Kim, Y.P.; Uchida, R.; Tomoda, H.; Omura, S. Taxonomy and secondary metabolites of Pseudobotrytis sp. FKA-25. Mycoscience 2004, 45, 9–16. [Google Scholar] [CrossRef]
- Kendall, J.K.; Fisher, T.H. An improved synthesis of 6,8-dimethoxy-3-methylisocoumarin, a fungal metabolite precursor. J. Org. Chem. 1989, 54, 4218–4220. [Google Scholar] [CrossRef]
- Cheng, M.J.; Wu, M.D.; Chen, I.S.; Chen, C.Y.; Lo, W.L.; Yuan, G.F. Secondary metabolites from the red mould rice of Monascus purpureus BCRC 38113. Nat. Prod. Res. 2010, 24, 1719–1725. [Google Scholar] [CrossRef] [PubMed]
- Pedras, M.S.C.; Taylor, J.L.; Morales, V.M. Phomaligin A and other yellow pigments in Phoma lingam and P. wasabiae. Phytochemistry 1995, 38, 1215–1222. [Google Scholar] [CrossRef]
- Elbandy, M.; Shinde, P.B.; Hong, J.; Bae, K.S.; Kim, M.A.; Lee, S.M.; Jung, J.H. α-Pyrones and yellow pigments from the sponge-derived fungus Paecilomyces lilacinus. Bull. Korean Chem. Soc. 2009, 30, 188–192. [Google Scholar]
- Wu, M.D.; Cheng, M.J.; Liu, T.W.; Chen, Y.L.; Chan, H.Y.; Chen, H.P.; Wu, W.J.; Chen, K.P.; Yuan, G.F. Chemical constituents of the fungus Monascus pilosus BCRC 38093-fermented rice. Chem. Nat. Compd. 2015, 51, 554–556. [Google Scholar] [CrossRef]
- Zin, W.W.M.; Buttachon, S.; Dethoup, T.; Pereira, J.A.; Gales, L.; Inacio, A.; Costa, P.M.; Lee, M.; Sekeroglu, N.; Silva, A.M.S.; et al. Antibacterial and antibiofilm activities of the metabolites isolated from the culture of the mangrove-derived endophytic fungus Eurotium chevalieri KUFA 0006. Phytochemistry 2017, 141, 86–97. [Google Scholar] [CrossRef]
- Chiu, H.W.; Fang, W.H.; Chen, Y.L.; Wu, M.D.; Yuan, G.F.; Ho, S.Y.; Wang, Y.J. Monascuspiloin enhances the radiation sensitivity of human prostate cancer cells by stimulating endoplasmic reticulum stress and inducing autophagy. PLoS ONE 2012, 7, e40462. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.J.; Hung, C.M.; Chen, Y.L.; Wu, M.D.; Yuan, G.F.; Wang, Y.J. Monascuspiloin induces apoptosis and autophagic cell death in human prostate cancer cells via the Akt and AMPK signaling pathways. J. Agric. Food Chem. 2012, 60, 7185–7193. [Google Scholar] [CrossRef]
- Yang, F.; Chen, W.D.; Deng, R.; Zhang, H.; Tang, J.; Wu, K.W.; Li, D.D.; Feng, G.K.; Lan, W.J.; Li, H.J.; et al. Hirsutanol A, a novel sesquiterpene compound from fungus Chondrostereum sp., induces apoptosis and inhibits tumor growth through mitochondrial-independent ROS production: Hirsutanol A inhibits tumor growth through ROS production. J. Transl. Med. 2013, 11, 32. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Chen, W.D.; Deng, R.; Li, D.D.; Wu, K.W.; Feng, G.K.; Li, H.J.; Zhu, X.F. Hirsutanol A induces apoptosis and autophagy via reactive oxygen species accumulation in breast cancer MCF-7 cells. J. Pharmacol. Sci. 2012, 119, 214–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Position | 1 a | 3 b | 4 b | |||
|---|---|---|---|---|---|---|
| δC, type | δH, mult. (J, Hz) | δC, type | δH, mult. (J, Hz) | δC, type | δH, mult. (J, Hz) | |
| 1-NH | 10.83, brs | 11.53, brs | 11.50, brs | |||
| 2 | 156.9, C | 157.0, C | 156.5, C | |||
| 3 | 159.5, C | 157.6, C | 161.7, C | |||
| 5 | 120.9, CH | 7.41, s | 120.0, CH | 7.30, s | 120.0, CH | 7.32, s |
| 6 | 135.1, C | 142.5, C | 142.0, C | |||
| 7 | 41.8, CH2 | 2.69, d (7.0) | 41.5, CH2 | 2.68, dd (14.4, 7.6) 2.62, dd (14.4, 7.2) | 36.6, CH | 3.24, ddq (6.8, 6.8, 6.8) |
| 8 | 26.9, CH | 2.23, tqq (7.0, 7.0, 7.0) | 26.9, CH | 2.18, m | 27.6, CH2 | 1.53, ddq (13.6, 6.8, 6.8) 1.79, ddq (13.6, 6.8, 6.8) |
| 9 | 22.7, CH3 | 0.98, d (7.0) | 22.6, CH3 | 0.95, d (6.6) | 11.9, CH3 | 0.89, t (8.4) |
| 10 | 22.7, CH3 | 0.98, d (7.0) | 22.6, CH3 | 0.95, d (6.6) | 17.6, CH3 | 1.21, d (6.8) |
| 11 | 140.8, C | 72.1, C | 72.2, C | |||
| 12 | 26.0, CH2 | 2.46, q (7.0) | 35.4, CH2 | 1.86, m | 35.3, CH2 | 1.87, m |
| 13 | 12.6, CH3 | 1.17, t (7.0) | 8.1, CH3 | 0.90, t (7.2) | 8.1, CH3 | 0.92, t (7.6) |
| 14 | 115.0, CH2 | a. 5.35, s b. 5.66, s | 27.1, CH3 | 1.57, s | 27.1, CH3 | 1.57, s |
| 11-OH | 3.77, brs | 3.482, brs | ||||
| 6 a | 13 b | ||||
|---|---|---|---|---|---|
| Position | δC, type | δH, mult. (J, Hz) | Position | δC, type | δH, mult. (J, Hz) |
| 1 | 166.5, C | 1 | 63.6, CH2 | a. 4.37, brd (16.2) b. 4.44, brd (16.2) | |
| 3 | 153.8, C | 3 | 73.4, CH | 4.00, m | |
| 4 | 106.3, CH | 6.30, s | 4 | 36.2, CH2 | a. 2.22, ddd (18.0, 3.0, 3.0) b. 2.35, dd (18.0, 9.0) |
| 4a | 136.5, C | 5 | 34.0, CH2 | 2.58, m | |
| 5 | 97.2, CH | 6.32, s | 6 | 41.3, CH | 3.04, ddd (13.2, 9.6, 6.0) |
| 6 | 164.6, C | 7 | 82.9, C | ||
| 7 | 112.6, C | 8 | 192.4, C | ||
| 8 | 159.9, C | 9 | 151.6, C | ||
| 8a | 99.9, C | 10 | 129.6, C | ||
| 9 | 43.0, CH2 | 2.59, dd (14.4, 8.0) 2.66, dd (14.4, 4.4) | 11 | 48.8, CH | 2.75, dd (13.2, 3.0) |
| 10 | 65.5, CH | 4.26, m | 12 | 174.7, C | |
| 11 | 23.2, CH3 | 1.30, d (6.0) | 13 | 130.0, CH | 5.52, ddq (15.6, 6.6, 1.2) |
| 12 | 7.9, CH3 | 2.12, s | 14 | 129.0, CH | 5.78, dqd (15.6, 6.6, 1.2) |
| 13 | 55.8, CH3 | 3.90, s | 15 | 17.8, CH3 | 1.73, ddd (6.6, 1.2, 0.6) |
| 8-OH | 11.15, brs | 16 | 16.6, CH3 | 1.42, s | |
| 10-OH | 1.71, brs | 17 | 69.4, CH | 4.22, m | |
| 18 | 35.0, CH2 | 1.55, m | |||
| 19 | 25.8, CH2 | 1.53, m 1.33, m | |||
| 20 | 31.5, CH2 | 1.33, m | |||
| 21 | 22.5, CH2 | 1.33, m | |||
| 22 | 14.0, CH3 | 0.91, t (7.2) | |||
| 17-OH | 2.07, d (4.8) | ||||
| Compounds | Human Nasopharyngeal Carcinoma Cell Lines | Human Hepatocellular Cancer Cell Lines | ||||
|---|---|---|---|---|---|---|
| CNE1 | CNE2 | HONE1 | SUNE1 | HepG2 | QGY7701 | |
| 1 | − a | − | − | 90.55 ± 1.58 | − | − |
| 2 | − | − | − | 92.53 ± 1.10 | − | − |
| 3 | 81.91 ±1.81 | − | − | − | − | − |
| 4 | 63.88 ± 1.22 | − | − | 92.78 ± 1.73 | − | − |
| 5 | − | 91.78 ± 1.90 | − | 64.35 ± 0.89 | − | − |
| 6 | − | 75.70 ± 1.09 | − | 72.07 ± 0.65 | 1.72 ± 0.35 | 0.71 ± 0.12 |
| 7 | − | 85.66 ± 1.69 | − | 28.12 ± 0.75 | 9.60 ± 0.46 | 7.12 ± 0.36 |
| 8 | − | − | − | 39.38 ± 0.58 | 46.10 ± 0.91 | 31.62 ± 1.23 |
| 9 | 70.96 ± 1.51 | − | − | − | − | − |
| 10 | 72.72 ± 1.36 | − | − | − | − | − |
| 11 | 92.87 ± 2.10 | − | − | − | − | − |
| 12 | − | − | − | − | − | − |
| 13 | 50.55 ± 0.88 | − | − | − | − | − |
| Hirsutanol A | 10.08 ± 0.92 | 12.72 ± 0.86 | 17.40 ± 0.52 | 3.50 ± 0.28 | 10.11 ± 0.69 | 21.12 ± 1.01 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ran, Y.-Q.; Lan, W.-J.; Qiu, Y.; Guo, Q.; Feng, G.-K.; Deng, R.; Zhu, X.-F.; Li, H.-J.; Dong, J. Monarubins A–C from the Marine Shellfish-Associated Fungus Monascus ruber BB5. Mar. Drugs 2020, 18, 100. https://doi.org/10.3390/md18020100
Ran Y-Q, Lan W-J, Qiu Y, Guo Q, Feng G-K, Deng R, Zhu X-F, Li H-J, Dong J. Monarubins A–C from the Marine Shellfish-Associated Fungus Monascus ruber BB5. Marine Drugs. 2020; 18(2):100. https://doi.org/10.3390/md18020100
Chicago/Turabian StyleRan, Yan-Qin, Wen-Jian Lan, Yi Qiu, Qi Guo, Gong-Kan Feng, Rong Deng, Xiao-Feng Zhu, Hou-Jin Li, and Jun Dong. 2020. "Monarubins A–C from the Marine Shellfish-Associated Fungus Monascus ruber BB5" Marine Drugs 18, no. 2: 100. https://doi.org/10.3390/md18020100