Characterization of the Shape Anisotropy of Superparamagnetic Iron Oxide Nanoparticles during Thermal Decomposition
Abstract
:1. Introduction
2. Materials and Methods
2.1. SPIONs Synthesis
2.2. Electron Tomography
2.3. Image Processing
2.4. Stereology, Sphericity and Anisotropy
2.5. Statistics
2.6. Small-Angle X-Ray Scattering
3. Results
3.1. Particle Volume
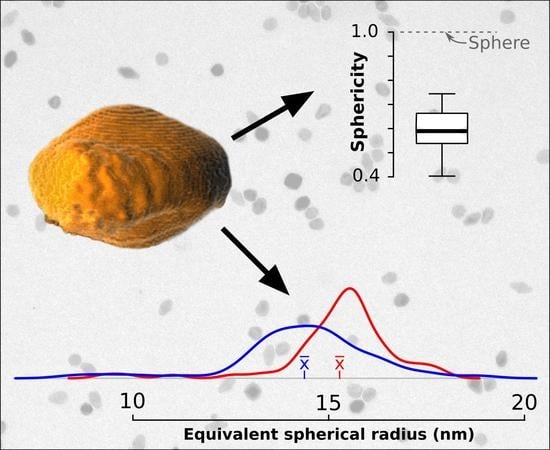
3.2. Sphericity, Anisotropy and Preferred Orientation
4. Discussion
4.1. The Thermal Decomposition Process
4.2. Measurement Disagreements
4.3. Model-Based Versus Model-Free Quantifcation
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A





References
- Bazylinski, D.A.; Frankel, R.B. Magnetosome formation in prokaryotes. Nat. Rev. Microbiol. 2004, 2, 217–230. [Google Scholar] [CrossRef] [Green Version]
- Lower, B.H.; Bazylinski, D.A. The Bacterial Magnetosome: A Unique Prokaryotic Organelle. J. Mol. Microbiol. Biotechnol. 2013, 23, 63–80. [Google Scholar] [CrossRef] [PubMed]
- Keim, C.N.; Martins, J.L.; Abreu, F.; Rosado, A.S.; Barros, H.L.; Borojevic, R.; Lins, U.; Farina, M. Multicellular life cycle of magnetotactic prokaryotes. FEMS Microbiol. Lett. 2004, 240, 203–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, F.F.; Yang, W.; Jiang, W.; Geng, S.; Peng, T.; Li, J.L. Magnetosomes eliminate intracellular reactive oxygen species in Magnetospirillum gryphiswaldense MSR-1. Environ. Microbiol. 2012, 14, 1722–1729. [Google Scholar] [CrossRef] [PubMed]
- Vincenti, B.; Ramos, G.; Cordero, M.L.; Douarche, C.; Soto, R.; Clément, E. Magnetotactic bacteria in a droplet self-assemble into a rotary motor. Nat. Commun. 2019, 10, 5082–5088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frankel, R.B.; Blakemore, R.P.; Wolfe, R.S. Magnetite in Freshwater Magnetotactic Bacteria. Science 1979, 203, 1355–1356. [Google Scholar] [CrossRef] [Green Version]
- Alphandéry, E. Applications of Magnetosomes Synthesized by Magnetotactic Bacteria in Medicine. Front. Bioeng. Biotechnol. 2014, 2. [Google Scholar] [CrossRef]
- Katzmann, E.; Eibauer, M.; Lin, W.; Pan, Y.; Plitzko, J.; Schüler, D. Analysis of Magnetosome Chains in Magnetotactic Bacteria by Magnetic Measurements and Automated Image Analysis of Electron Micrographs. Appl. Environ. Microbiol. 2013, 79, 7755–7762. [Google Scholar] [CrossRef] [Green Version]
- Bazylinski, D.A.; Garratt-Reed, A.J.; Frankel, R.B. Electron microscopic studies of magnetosomes in magnetotactic bacteria. Microsc. Res. Tech. 1994, 27, 389–401. [Google Scholar] [CrossRef]
- Scheffel, A.; Gärdes, A.; Grünberg, K.; Wanner, G.; Schüler, D. The Major Magnetosome Proteins MamGFDC Are Not Essential for Magnetite Biomineralization in Magnetospirillum gryphiswaldense but Regulate the Size of Magnetosome Crystals. J. Bacteriol. 2007, 190, 377–386. [Google Scholar] [CrossRef] [Green Version]
- Murat, D.; Quinlan, A.; Vali, H.; Komeili, A. Comprehensive genetic dissection of the magnetosome gene island reveals the step-wise assembly of a prokaryotic organelle. Proc. Natl. Acad. Sci. USA 2010, 107, 5593–5598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahn-Lee, L.; Komeili, A. The magnetosome model: Insights into the mechanisms of bacterial biomineralization. Front. Microbiol. 2013, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Revia, R.A.; Zhang, M. Magnetite nanoparticles for cancer diagnosis, treatment, and treatment monitoring: Recent advances. Mater. Today 2016, 19, 157–168. [Google Scholar] [CrossRef]
- Kuchma, E.; Zolotukhin, P.V.; Belanova, A.; Soldatov, M.; Lastovina, T.; Kubrin, S.; Nikolsky, A.V.; Mirmikova, L.; Soldatov, A.V. Low toxic maghemite nanoparticles for theranostic applications. Int. J. Nanomed. 2017, 12, 6365–6371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wahajuddin, M.; Arora, S. Superparamagnetic iron oxide nanoparticles: Magnetic nanoplatforms as drug carriers. Int. J. Nanomed. 2012, 7, 3445–3471. [Google Scholar] [CrossRef] [Green Version]
- Mahmoudi, M.; Hofmann, H.; Rothen-Rutishauser, B.; Petri-Fink, A. Assessing the In Vitro and In Vivo Toxicity of Superparamagnetic Iron Oxide Nanoparticles. Chem. Rev. 2011, 112, 2323–2338. [Google Scholar] [CrossRef] [Green Version]
- Petri-Fink, A.; Steitz, B.; Finka, A.; Salaklang, J.; Hofmann, H. Effect of cell media on polymer coated superparamagnetic iron oxide nanoparticles (SPIONs): Colloidal stability, cytotoxicity, and cellular uptake studies. Eur. J. Pharm. Biopharm. 2008, 68, 129–137. [Google Scholar] [CrossRef]
- Khalkhali, M.; Rostamizadeh, K.; Sadighian, S.; Khoeini, F.; Naghibi, M.; Hamidi, M. The impact of polymer coatings on magnetite nanoparticles performance as MRI contrast agents: A comparative study. DARU J. Pharm. Sci. 2015, 23, 45. [Google Scholar] [CrossRef] [Green Version]
- Ko, S.; Huh, C. Use of nanoparticles for oil production applications. J. Pet. Sci. Eng. 2019, 172, 97–114. [Google Scholar] [CrossRef]
- Manaenkov, O.V.; Matveeva, V.G.; Sinitzyna, P.V.; Ratkevich, E.A.; Kislitza, O.V.; Doluda, V.Y.; Sulman, E.M.; Sidorov, A.I.; Mann, J.J.; Losovyj, Y.; et al. Magnetically recoverable catalysts for cellulose conversion into glycols. Chem. Eng. Trans. 2016, 52, 637–642. [Google Scholar] [CrossRef]
- Digigow, R.G.; Dechézelles, J.-F.; Kaufmann, J.; Vanhecke, D.; Knapp, H.; Lattuada, M.; Rothen-Rutishauser, B.; Petri-Fink, A. Magnetic microreactors for efficient and reliable magnetic nanoparticle surface functionalization. Lab Chip 2014, 14, 2276–2286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nano-Safety: What We Need to Know to Protect Workers; Fazarro, D. (Ed.) De Gruyter textbook; De Gruyter: Berlin, Germany; Boston, MA, USA, 2017; ISBN 978-3-11-037375-2. [Google Scholar]
- Stephen, Z.R.; Kievit, F.M.; Zhang, M. Magnetite nanoparticles for medical MR imaging. Mater. Today 2011, 14, 330–338. [Google Scholar] [CrossRef]
- Dadfar, S.M.; Camozzi, D.; Darguzyte, M.; Roemhild, K.; Varvarà, P.; Metselaar, J.; Banala, S.; Straub, M.; Güvener, N.; Engelmann, U.; et al. Size-isolation of superparamagnetic iron oxide nanoparticles improves MRI, MPI and hyperthermia performance. J. Nanobiotechnol. 2020, 18, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Barrow, M.; Taylor, A.; Fuentes-Caparrós, A.M.; Sharkey, J.; Daniels, L.M.; Mandal, P.; Park, B.K.; Murray, P.; Rosseinsky, M.J.; Adams, D.J. SPIONs for cell labelling and tracking using MRI: Magnetite or maghemite? Biomater. Sci. 2017, 6, 101–106. [Google Scholar] [CrossRef] [Green Version]
- Jasmin; De Souza, G.T.; Louzada, R.A.; Rosado-De-Castro, P.H.; Mendez-Otero, R.; De Carvalho, A.C.C. Tracking stem cells with superparamagnetic iron oxide nanoparticles: Perspectives and considerations. Int. J. Nanomed. 2017, 12, 779–793. [Google Scholar] [CrossRef] [Green Version]
- Bonnaud, C.; Monnier, C.; Demurtas, D.; Jud, C.; Vanhecke, D.; Montet, X.; Hovius, R.; Lattuada, M.; Rothen-Rutishauser, B.; Petri-Fink, A. Insertion of Nanoparticle Clusters into Vesicle Bilayers. ACS Nano 2014, 8, 3451–3460. [Google Scholar] [CrossRef]
- Laurent, S.; Saei, A.A.; Behzadi, S.; Panahifar, A.; Mahmoudi, M. Superparamagnetic iron oxide nanoparticles for delivery of therapeutic agents: Opportunities and challenges. Expert Opin. Drug Deliv. 2014, 11, 1449–1470. [Google Scholar] [CrossRef]
- Laurent, S.; Dutz, S.; Häfeli, U.O.; Mahmoudi, M. Magnetic fluid hyperthermia: Focus on superparamagnetic iron oxide nanoparticles. Adv. Colloid Interface Sci. 2011, 166, 8–23. [Google Scholar] [CrossRef]
- Gamarra, L.F.; Silva, A.C.; De Oliveira, T.R.; Mamani, J.B.; Malheiros, S.M.; Malavolta, L.; Pavon, L.F.; Sibov, T.T.; Junior, E.A.; Amaro, E.; et al. Application of hyperthermia induced by superparamagnetic iron oxide nanoparticles in glioma treatment. Int. J. Nanomed. 2011, 6, 591–603. [Google Scholar] [CrossRef] [Green Version]
- Piazza, R.D.; Viali, W.; Dos Santos, C.C.; Nunes, E.S.; Marques, R.F.C.; De Morais, P.C.; Da Silva, S.W.; Coaquira, J.A.H.; Jafelicci, M.; Da Silva, E.N.; et al. PEGlatyon-SPION surface functionalization with folic acid for magnetic hyperthermia applications. Mater. Res. Express 2020, 7, 015078. [Google Scholar] [CrossRef]
- De Montferrand, C.; Hu, L.; Milosevic, I.; Russier, V.; Bonnin, D.; Motte, L.; Brioude, A.; Lalatonne, Y. Iron oxide nanoparticles with sizes, shapes and compositions resulting in different magnetization signatures as potential labels for multiparametric detection. Acta Biomater. 2013, 9, 6150–6157. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Yang, S.; Wu, W. Shape control of inorganic nanoparticles from solution. Nanoscale 2016, 8, 1237–1259. [Google Scholar] [CrossRef] [PubMed]
- Kinnear, C.; Moore, T.L.; Lorenzo, L.R.; Rothen-Rutishauser, B.; Petri-Fink, A. Form Follows Function: Nanoparticle Shape and Its Implications for Nanomedicine. Chem. Rev. 2017, 117, 11476–11521. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Zhu, X.; Wu, D.; Chen, Q.; Huang, D.; Sun, C.; Xin, J.; Ni, K.; Gao, J. Anisotropic Shaped Iron Oxide Nanostructures: Controlled Synthesis and Proton Relaxation Shortening Effects. Chem. Mater. 2015, 27, 3505–3515. [Google Scholar] [CrossRef]
- Andrade, R.G.D.; Veloso, S.R.S.; Castanheira, E.M.S. Shape Anisotropic Iron Oxide-Based Magnetic Nanoparticles: Synthesis and Biomedical Applications. Int. J. Mol. Sci. 2020, 21, 2455. [Google Scholar] [CrossRef] [Green Version]
- Luchini, A.; Heenan, R.K.; Paduano, L.; Vitiello, G. Functionalized SPIONs: The surfactant nature modulates the self-assembly and cluster formation. Phys. Chem. Chem. Phys. 2016, 18, 18441–18449. [Google Scholar] [CrossRef]
- Wetterskog, E.; Agthe, M.; Mayence, A.; Grins, J.; Wang, N.; Rana, S.; Ahniyaz, A.; Salazar-Alvarez, G.; Bergström, L. Precise control over shape and size of iron oxide nanocrystals suitable for assembly into ordered particle arrays. Sci. Technol. Adv. Mater. 2014, 15, 55010. [Google Scholar] [CrossRef]
- Ganguly, A.; Kundu, R.; Ramanujachary, K.; Lofland, S.; Das, D.; Vasanthacharya, N.Y.; Ahmad, T.; Ganguli, A. Role of carboxylate ion and metal oxidation state on the morphology and magnetic properties of nanostructured metal carboxylates and their decomposition products. J. Chem. Sci. 2008, 120, 521–528. [Google Scholar] [CrossRef]
- Simeonidis, K.; Morales, M.P.; Marciello, M.; Angelakeris, M.; De La Presa, P.; Lazaro-Carrillo, A.; Tabero, A.; Villanueva, A.; Chubykalo-Fesenko, O.; Serantes, D. In-situ particles reorientation during magnetic hyperthermia application: Shape matters twice. Sci. Rep. 2016, 6, 38382. [Google Scholar] [CrossRef] [Green Version]
- Usov, N.; Nesmeyanov, M.S.; Tarasov, V.P. Magnetic Vortices as Efficient Nano Heaters in Magnetic Nanoparticle Hyperthermia. Sci. Rep. 2018, 8, 1224. [Google Scholar] [CrossRef]
- Ramzannezhad, A.; Gill, P.; Bahari, A. Fabrication of magnetic nanorods and their applications in medicine. BioNanoMaterials 2017, 18. [Google Scholar] [CrossRef]
- Geng, S.; Yang, H.; Ren, X.; Liu, Y.; He, S.; Zhou, J.; Su, N.; Li, Y.; Xu, C.; Zhang, X.; et al. Anisotropic Magnetite Nanorods for Enhanced Magnetic Hyperthermia. Chem. Asian J. 2016, 11, 2996–3000. [Google Scholar] [CrossRef] [PubMed]
- Kovalenko, M.V.; Bodnarchuk, M.I.; Lechner, R.T.; Hesser, G.; Schäffler, F.; Heiss, W. Fatty Acid Salts as Stabilizers in Size- and Shape-Controlled Nanocrystal Synthesis: The Case of Inverse Spinel Iron Oxide. J. Am. Chem. Soc. 2007, 129, 6352–6353. [Google Scholar] [CrossRef] [PubMed]
- Hufschmid, R.; Arami, H.; Ferguson, R.M.; Gonzales, M.; Teeman, E.; Brush, L.N.; Browning, N.D.; Krishnan, K.M. Synthesis of phase-pure and monodisperse iron oxide nanoparticles by thermal decomposition. Nanoscale 2015, 7, 11142–11154. [Google Scholar] [CrossRef] [Green Version]
- Prijic, S.; Scancar, J.; Romih, R.; Cemazar, M.; Bregar, V.B.; Žnidaršič, A.; Sersa, G. Increased Cellular Uptake of Biocompatible Superparamagnetic Iron Oxide Nanoparticles into Malignant Cells by an External Magnetic Field. J. Membr. Boil. 2010, 236, 167–179. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Senesi, A.J.; Lee, B. Small Angle X-ray Scattering for Nanoparticle Research. Chem. Rev. 2016, 116, 11128–11180. [Google Scholar] [CrossRef]
- Park, J.; An, K.; Hwang, Y.; Park, J.-G.; Noh, H.-J.; Kim, J.-Y.; Park, J.-H.; Hwang, N.-M.; Hyeon, T. Ultra-large-scale syntheses of monodisperse nanocrystals. Nat. Mater. 2004, 3, 891–895. [Google Scholar] [CrossRef]
- Howard, V.; Reed, M. Unbiased Stereology: Three-Dimensional Measurement in Microscopy, 2nd ed.; Advanced Methods; Garland Science: New York, NY, USA, 2005. [Google Scholar]
- Gundersen, H.J.G.; Jensen, E.B. The efficiency of systematic sampling in stereology and its prediction. J. Microsc. 1987, 147, 229–263. [Google Scholar] [CrossRef]
- Mastronarde, D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Boil. 2005, 152, 36–51. [Google Scholar] [CrossRef]
- Kremer, J.R.; Mastronarde, D.N.; McIntosh, J. Computer Visualization of Three-Dimensional Image Data Using IMOD. J. Struct. Boil. 1996, 116, 71–76. [Google Scholar] [CrossRef] [Green Version]
- Frank, J. Electron Tomography: Three-Dimensional Imaging with the Transmission Electron Microscope; Plenum Press: New York, NY, USA, 1992. [Google Scholar]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jennings, B.R.; Parslow, K. Particle Size Measurement: The Equivalent Spherical Diameter. Proc. R. Soc. Lond. A 1988, 419, 137–149. [Google Scholar] [CrossRef]
- Bolte, S.; Cordelières, F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006, 224, 213–232. [Google Scholar] [CrossRef] [PubMed]
- Collins, T.J. ImageJ for microscopy. Biotechniques 2007, 43, S25–S30. [Google Scholar] [CrossRef] [PubMed]
- Vanhecke, D.; Lorenzo, L.R.; Kinnear, C.; Durantie, E.; Rothen-Rutishauser, B.; Petri-Fink, A. Assumption-free morphological quantification of single anisotropic nanoparticles and aggregates. Nanoscale 2017, 9, 4918–4927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michel, R.P.; Cruz-Orive, L. Application of the Cavalieri principle and vertical sections method to lung: Estimation of volume and pleural surface area. J. Microsc. 1988, 150, 117–136. [Google Scholar] [CrossRef] [PubMed]
- Kubinova, L.; Janáček, J. Estimating surface area by the isotropic fakir method from thick slices cut in an arbitrary direction. J. Microsc. 1998, 191, 201–211. [Google Scholar] [CrossRef]
- Wadell, H. Volume, Shape, and Roundness of Quartz Particles. J. Geol. 1935, 43, 250–280. [Google Scholar] [CrossRef]
- Basser, P.J.; Pierpaoli, C. Microstructural and Physiological Features of Tissues Elucidated by Quantitative-Diffusion-Tensor MRI. J. Magn. Reson. Ser. B 1996, 111, 209–219. [Google Scholar] [CrossRef]
- Meijering, E. TransformJ: ImageJ plugin suite for geometrical image transformation. Available online: https://github.com/imagescience/TransformJ/ (accessed on 15 March 2020).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Grillo, I. Small-Angle Neutron Scattering and Applications in Soft Condensed Matter. In Soft Matter Characterization; Borsali, R., Pecora, R., Eds.; Springer: Dordrecht, The Netherlands, 2008; pp. 723–782. ISBN 978-1-4020-4464-9. [Google Scholar]
- Pauw, B.R. Everything SAXS: Small-angle scattering pattern collection and correction. J. Phys. Condens. Matter 2013, 25, 383201. [Google Scholar] [CrossRef]
- Crippa, F.; Rodriguez-Lorenzo, L.; Hua, X.; Goris, B.; Bals, S.; Garitaonandia, J.S.; Balog, S.; Burnand, D.; Hirt, A.M.; Haeni, L.; et al. Phase Transformation of Superparamagnetic Iron Oxide Nanoparticles via Thermal Annealing: Implications for Hyperthermia Applications. ACS Appl. Nano Mater. 2019, 2, 4462–4470. [Google Scholar] [CrossRef] [Green Version]
- Lee, N.; Yoo, N.; Ling, D.; Cho, M.H.; Hyeon, T.; Cheon, J. Iron Oxide Based Nanoparticles for Multimodal Imaging and Magnetoresponsive Therapy. Chem. Rev. 2015, 115, 10637–10689. [Google Scholar] [CrossRef] [PubMed]
- Bonnaud, C.; Vanhecke, D.; Demurtas, D.; Rothen-Rutishauser, B.; Petri-Fink, A. Spatial SPION Localization in Liposome Membranes. IEEE Trans. Magn. 2012, 49, 166–171. [Google Scholar] [CrossRef] [Green Version]
- Prakash, Y.S.; Smithson, K.G.; Sieck, G.C. Application of the Cavalieri Principle in Volume Estimation Using Laser Confocal Microscopy. NeuroImage 1994, 1, 325–333. [Google Scholar] [CrossRef]
- Vanhecke, D.; Studer, D.; Ochs, M. Stereology meets electron tomography: Towards quantitative 3D electron microscopy. J. Struct. Boil. 2007, 159, 443–450. [Google Scholar] [CrossRef]
- Thanh, N.T.K.; MacLean, N.; Mahiddine, S. Mechanisms of Nucleation and Growth of Nanoparticles in Solution. Chem. Rev. 2014, 114, 7610–7630. [Google Scholar] [CrossRef]
- Xia, Y.; Xiong, Y.; Lim, B.; Skrabalak, S.E. Shape-Controlled Synthesis of Metal Nanocrystals: Simple Chemistry Meets Complex Physics? Angew. Chem. Int. Ed. 2009, 48, 60–103. [Google Scholar] [CrossRef]
- Rottman, C.; Wortis, M. Equilibrium crystal shapes for lattice models with nearest-and next-nearest-neighbor interactions. Phys. Rev. B 1984, 29, 328–339. [Google Scholar] [CrossRef]
- Kwon, S.G.; Piao, Y.; Park, J.; Angappane, S.; Jo, Y.; Hwang, N.-M.; Park, J.-G.; Hyeon, T. Kinetics of Monodisperse Iron Oxide Nanocrystal Formation by “Heating-Up” Process. J. Am. Chem. Soc. 2007, 129, 12571–12584. [Google Scholar] [CrossRef]
- Midgley, P.A.; Dunin-Borkowski, R.E. Electron tomography and holography in materials science. Nat. Mater. 2009, 8, 271–280. [Google Scholar] [CrossRef]
- Glaeser, R.M. How good can cryo-EM become? Nat. Methods 2015, 13, 28–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glaeser, R.M.; Han, B.-G. Opinion: Hazards faced by macromolecules when confined to thin aqueous films. Biophys. Rep. 2016, 3, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, Y.Z.; Baldwin, P.R.; Davis, J.H.; Williamson, J.R.; Potter, C.; Carragher, B.; Lyumkis, D. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods 2017, 14, 793–796. [Google Scholar] [CrossRef] [PubMed]
- Jordan, A.; Wust, P.; Fählin, H.; John, W.; Hinz, A.; Felix, R. Inductive heating of ferrimagnetic particles and magnetic fluids: Physical evaluation of their potential for hyperthermia. Int. J. Hyperth. 1993, 9, 51–68. [Google Scholar] [CrossRef]
- Rosensweig, R. Heating magnetic fluid with alternating magnetic field. J. Magn. Magn. Mater. 2002, 252, 370–374. [Google Scholar] [CrossRef]
- Lorenzo, L.R.; Alvarez-Puebla, R.A.; Pastoriza-Santos, I.; Mazzucco, S.; Steéphan, O.; Kociak, M.; Liz-Marzán, L.M.; De Abajo, F.J.G. Zeptomol Detection Through Controlled Ultrasensitive Surface-Enhanced Raman Scattering. J. Am. Chem. Soc. 2009, 131, 4616–4618. [Google Scholar] [CrossRef]





| Measure | Onset | 10′ | 20′ | 30′ | Cooldown |
|---|---|---|---|---|---|
| Particle count | 149 | 112 | 83 | 96 | 72 |
| Volume (nm3) | |||||
| Mean | 1597 ± 1040 | 12,085 ± 3938 | 14,325 ± 2981 | 16,239 ± 3849 | 15,324 ± 3732 |
| Median | 1470 | 11,341 | 13,990 | 16,052 | 15,400 |
| Surface area (nm2) | |||||
| Mean | 679 ± 287 | 3528 ± 1471 | 3746 ± 927 | 7700 ± 1766 | 5144 ± 1490 |
| Median | 642 | 3124 | 3559 | 5383 | 5019 |
| Shape Factors | |||||
| Sphericity | 0.91 ± 0.09 | 0.76 ± 0.12 | 0.77 ± 0.10 | 0.56 ± 0.09 | 0.59 ± 0.08 |
| Anisotropy | 0.047 ± 0.027 | 0.058 ± 0.031 | 0.062 ± 0.039 | 0.0831 ± 0.08 | 0.082 ± 0.040 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vanhecke, D.; Crippa, F.; Lattuada, M.; Balog, S.; Rothen-Rutishauser, B.; Petri-Fink, A. Characterization of the Shape Anisotropy of Superparamagnetic Iron Oxide Nanoparticles during Thermal Decomposition. Materials 2020, 13, 2018. https://doi.org/10.3390/ma13092018
Vanhecke D, Crippa F, Lattuada M, Balog S, Rothen-Rutishauser B, Petri-Fink A. Characterization of the Shape Anisotropy of Superparamagnetic Iron Oxide Nanoparticles during Thermal Decomposition. Materials. 2020; 13(9):2018. https://doi.org/10.3390/ma13092018
Chicago/Turabian StyleVanhecke, Dimitri, Federica Crippa, Marco Lattuada, Sandor Balog, Barbara Rothen-Rutishauser, and Alke Petri-Fink. 2020. "Characterization of the Shape Anisotropy of Superparamagnetic Iron Oxide Nanoparticles during Thermal Decomposition" Materials 13, no. 9: 2018. https://doi.org/10.3390/ma13092018