Electrochemical Detection of Solution Phase Hybridization Related to Single Nucleotide Mutation by Carbon Nanofibers Enriched Electrodes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Apparatus and Chemicals
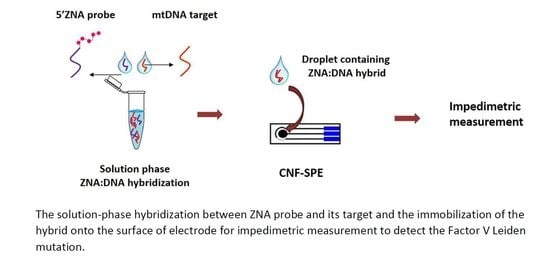
2.2. Procedure
- (i)
- Hybridization of the ZNA probe with the mDNA target, or wDNA, non-complementary oligonucleotides (C-DNA, T-DNA, NC-1, NC-2), mutant type PCR products; mPCR-1 and mPCR-2 and wild type PCR products; wPCR-1 and wPCR-2 in the solution phase.
- (ii)
- Immobilization of the hybrid of ZNA:DNA as well as others onto the surface of CNF-SPEs.
- (iii)
- Measurements via electrochemical impedance spectroscopy (EIS) technique.
2.3. Impedimetric Measurements
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pellestor, F.; Paulasova, P. The peptide nucleic acids, efficient tools for molecular diagnosis (Review). Int. J. Mol. Med. 2004, 132, 521–525. [Google Scholar] [CrossRef]
- Pons, B.; Kotera, M.; Zuber, G.; Behr, J.P. Online synthesis of diblock cationic oligonucleotides for enhanced hybridization to their complementary sequence. Chembiochem 2006, 7, 1173–1176. [Google Scholar] [CrossRef] [PubMed]
- Moreau, V.; Voirin, E.; Paris, C.; Kotera, M.; Nothisen, M.; Remy, J.S.; Behr, J.P.; Erbacher, P.; Lenne-Samuel, N. Zip Nucleic Acids: New high affinity oligonucleotides as potent primers for PCR and reverse transcription. Nucleic Acids Res. 2009, 37, e130. [Google Scholar] [CrossRef] [PubMed]
- Noir, R.; Kotera, M.; Pons, B.; Remy, J.S.; Behr, J.P. Oligonucleotide- Oligospermine Conjugates (Zip Nucleic Acids): A Convenient Means of Finely Tuning Hybridization Temperatures. J. Am. Chem. Soc. 2008, 9, 13500–13505. [Google Scholar] [CrossRef]
- Kypr, J.; Kejnovska, I.; Renciuk, D.; Vorlickova, M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009, 37, 1713–1725. [Google Scholar] [CrossRef] [Green Version]
- Bi, S.; Yue, S.; Zhang, S. Hybridization chain reaction: A versatile molecular tool for biosensing, bioimaging, and biomedicine. Chem. Soc. Rev. 2017, 46, 4281–4298. [Google Scholar] [CrossRef]
- Khafizov, K.; Ivanov, M.V.; Glazova, O.V.; Kovalenko, S.P. Computational approaches to study the effects of small genomic variations. J. Mol. Model. 2015, 21, 251–265. [Google Scholar] [CrossRef]
- Erdem, A.; Congur, G. Dendrimer modified 8-channel screen-printed electrochemical array system for impedimetric detection of activated protein C. Sens. Actuators B Chem. 2014, 196, 168–174. [Google Scholar] [CrossRef]
- Chowdhury, A.D.; Ganganboina, A.B.; Park, E.Y.; Doong, R. Impedimetric biosensor for detection of cancer cells employing carbohydrate targeting ability of Concanavalin A. Biosens. Bioelectron. 2018, 122, 95–103. [Google Scholar] [CrossRef]
- Lee, H.; Keem, J.O.; Cho, H.; Choi, J.M.; Chung, W.S.; Jeon, D.Y.; Lee, D.S.; Shin, Y.B. High-performance nanogap electrode-based impedimetric sensor for direct DNA assays. Biosens. Bioelectron. 2018, 118, 153–159. [Google Scholar] [CrossRef]
- Erdem, A.; Congur, G. Impedimetric detection of in situ interaction between anti-cancer drug bleomycin and DNA. Int. J. Biol. Macromol. 2013, 61, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Top, M.; Er, O.; Congur, G.; Erdem, A.; Lambrecht Yurt, F. Intracellular uptake study of radiolabeled anticancer drug and impedimetric detection of its interaction with DNA. Talanta 2016, 160, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Congur, G.; Eksin, E.; Erdem, A. Impedimetric Detection of microRNA at Graphene Oxide Modified Sensors. Electrochim Acta 2015, 172, 20–27. [Google Scholar] [CrossRef]
- Pänke, O.; Balkenhohl, T.; Kafka, J.; Schäfer, D.; Lisdat, F. Impedance Spectroscopy and Biosensing. In Advances in Biochemical Engineering/Biotechnology; Scheper, T., Ed.; Springer: Berlin/Heidelberg, Germany, 2008; Volume 109, pp. 195–238. [Google Scholar]
- Li, P.Q.; Piper, A.; Schmueser, I.; Mountb, A.R.; Corrigan, D.K. Impedimetric measurement of DNA–DNA hybridisation using microelectrodes with different radii for detection of methicillin resistant Staphylococcus aureus (MRSA). Analyst 2017, 142, 1946–1952. [Google Scholar] [CrossRef]
- Chin, Y.T.; Liao, E.C.; Wu, C.C.; Wang, G.J.; Tsai, J.J. Label-free detection of single-nucleotide polymorphisms associated with myeloid differentiation-2 using a nanostructured biosensor. Biosens. Bioelectron. 2013, 49, 506–511. [Google Scholar] [CrossRef]
- Fernandez, R.E.; Williams, S.E.; Li, R.; Zhou, A. Gapped-duplex structure to label-free mismatch detection of pathogen DNA on solid substrate. Electrochem. Commun. 2015, 56, 1–5. [Google Scholar] [CrossRef]
- Zheng, Q.; Wu, H.; Shen, Z.; Gao, W.; Yu, Y.; Ma, Y.; Guang, W.; Guo, Q.; Yan, R.; Wang, J.; et al. An electrochemical DNA sensor based on polyaniline/graphene: High sensitivity to DNA sequences in a wide range. Analyst 2015, 140, 6660–6670. [Google Scholar] [CrossRef]
- Rosendaal, F.R.; Reitsma, P.H. Genetics of venous thrombosis. J. Thromb. Haemost. 2009, 7, 301–304. [Google Scholar] [CrossRef]
- Kujovich, J.L. Factor V Leiden thrombophilia. Nature 2011, 13, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Ren, Y.; Rezania, S.; Kang, K.A. Biosensor for Diagnosing Factor V Leiden, A Single Amino Acid Mutated Abnormality of Factor V. Adv. Exp. Med. Biol. 2008, 614, 245–252. [Google Scholar]
- Vlachou, M.A.; Glynou, K.M.; Ioannou, P.C.; Christopoulos, T.K.; Vartholomatos, G. Development of a three-biosensor panel for the visual detection of thrombophilia-associated mutations. Biosens. Bioelectron. 2010, 26, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.A.; Ren, Y.; Sharma, V.R.; Peiper, S.C. Near real-time immuno-optical sensor for diagnosing single point mutation, A model system: Sensor for Factor V Leiden diagnosis. Biosens. Bioelectron. 2009, 24, 2785–2790. [Google Scholar] [CrossRef] [PubMed]
- Ozkan, D.; Erdem, A.; Kara, P.; Kerman, K.; Meric, B.; Hassmann, J.; Ozsoz, M. The Allele—Specific genotype detection of Factor V Leiden mutation from PCR amplicons based on label free electrochemical genosensor. Anal. Chem. 2002, 74, 5931–5936. [Google Scholar] [CrossRef] [PubMed]
- Ozsoz, M.; Erdem, A.; Kerman, K.; Ozkan, D.; Tugrul, B.; Topcuoglu, N.; Ekren, H.; Taylan, M. Electrochemical genosensor based on colloidal gold nanoparticle for the detection of the Factor V Leiden mutation using disposable pencil graphite electrodes. Anal. Chem. 2003, 75, 2181–2187. [Google Scholar] [CrossRef]
- Paris, C.; Moreau, V.; Deglane, G.; Voirin, E.; Erbacher, P.; Lenne-Samuel, N. Zip nucleic acids are potent hydrolysis probes for quantitative PCR. Nucleic Acids Res. 2010, 38, e95. [Google Scholar] [CrossRef]
- Alvandi, E.; Koohdani, F. Zip nucleic acid: A new reliable method to increase the melting temperature of real-time PCR probes. J. Diabetes Metab. Disord. 2014, 13, 26–30. [Google Scholar] [CrossRef]
- Cui, H.; Kalinin, S.V.; Yang, X.; Lowndes, D.H. Growth of Carbon Nanofibers on Tipless Cantilevers for High Resolution Topography and Magnetic Force Imaging. Nano Lett. 2004, 4, 2157–2161. [Google Scholar] [CrossRef]
- Lee, C.S.; Baker, S.E.; Marcus, M.S.; Yang, W.S.; Eriksson, M.A.; Hamers, R.J. Electrically Addressable Biomolecular Functionalization of Carbon Nanotube and Carbon Nanofiber Electrodes. Nano Lett. 2004, 4, 1713–1716. [Google Scholar] [CrossRef]
- Erdem, A.; Eksin, E.; Congur, G. Indicator-free electrochemical biosensor for microRNA detection based on carbon nanofibers modified screen printed electrodes. J. Electroanal. Chem. 2015, 755, 167–173. [Google Scholar] [CrossRef]
- Erdem, A.; Congur, G.; Mayer, G. Aptasensor platform based on carbon nanofibers enriched screen printed electrodes for impedimetric detection of thrombin. J. Electroanal. Chem. 2015, 758, 12–19. [Google Scholar] [CrossRef]
- Xu, Q.; Liu, Z.; Fu, J.; Zhao, W.; Guo, Y.; Sun, X.; Zhang, H. Ratiometric electrochemical aptasensor based on ferrocene and carbon nanofibers for highly specific detection of tetracycline residues. Sci. Rep. 2017, 7, 14729. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wang, X.; Sun, X.; Guo, Y. Multiplex electrochemical aptasensor for detecting multiple antibiotics residues based on carbon fiber and mesoporous carbon-gold nanoparticles. Sens. Actuators B Chem. 2018, 265, 217–226. [Google Scholar] [CrossRef]
- Söderlund, H. DNA hybridization: Comparison of liquid and solid phase formats. Ann. Biol. Clin. 1990, 48, 489–491. [Google Scholar]
- Liu, W.T.; Guo, H.; Wu, J.H. Effects of Target Length on the Hybridization Efficiency and Specificity of rRNA-Based Oligonucleotide Microarrays. Appl. Environ. Microbiol. 2007, 73, 73–82. [Google Scholar] [CrossRef] [Green Version]
- Erdem, A.; Eksin, E. Magnetic beads assay based on Zip nucleic acid for electrochemical detection of Factor V Leiden mutation. Int. J. Biol. Macromol. 2019, 125, 839–846. [Google Scholar] [CrossRef]
- Erdem, A.; Eksin, E. ZNA probe immobilized single-use electrodes for impedimetric detection of nucleic acid hybridization related to single nucleotide mutation. Anal. Chim. Acta 2019, 1071, 78–85. [Google Scholar] [CrossRef]
- Erdem, A.; Eksin, E. Zip nucleic acid based single-use biosensor for electrochemical detection of Factor V Leiden mutation. Sens. Actuators B Chem. 2019, 288, 634–640. [Google Scholar] [CrossRef]
- Cummings, T.E.; Elving, P.J. Determination of the electrochemically effective electrode area. Anal. Chem. 1978, 50, 480–488. [Google Scholar] [CrossRef]
- Salgado-Figueroa, P.; Jara-Ulloa, P.; Alvarez-Lueje, A.; Squella, J.A. Sensitive Determination of Nitrofurantoin by Flow Injection Analysis Using Carbon Nanofiber Screen Printed Electrodes. Electroanalysis 2013, 25, 1433–1438. [Google Scholar] [CrossRef]
- Siddiqui, S.; Arumugam, P.U.; Chen, H.; Li, J.; Meyyappan, M. Characterization of Carbon Nanofiber Electrode Arrays Using Electrochemical Impedance Spectroscopy: Effect of Scaling Down Electrode Size. ACS Nano 2010, 4, 955–961. [Google Scholar] [CrossRef]
- Yue, Y.; Hu, G.; Zheng, M.; Guo, Y.; Cao, J.; Shao, S. A mesoporous carbon nanofiber-modified pyrolytic graphite electrode used for the simultaneous determination of dopamine, uric acid, and ascorbic acid. Carbon 2012, 50, 107–114. [Google Scholar] [CrossRef]
- Arumugam, P.U.; Chen, H.; Siddiqui, S.; Weinrich, J.A.P.; Jejelowod, A.; Li, J.; Meyyappan, M. Wafer-scale fabrication of patterned carbon nanofiber nanoelectrode arrays: A route for development of multiplexed, ultrasensitive disposable biosensors. Biosens. Bioelectron. 2009, 24, 2818–2824. [Google Scholar] [CrossRef] [PubMed]
- Eksin, E.; Erdem, A. Chitosan-carbon Nanofiber Modified Single-use Graphite Electrodes Developed for Electrochemical Detection of DNA Hybridization Related to Hepatitis B Virus. Electroanalysis 2016, 28, 2514–2521. [Google Scholar] [CrossRef]
- Mozo, J.D.; Carbajo, J.; Sturm, J.C.; Núñez-Vergara, L.; Salgado, P.; Squella, J.A. Determination of Nifuroxazide by Flow Injection Linear Adsorptive Stripping Voltammetry on a Screen-Printed Carbon Nanofiber Modified Electrode. Electroanalysis 2012, 24, 676–682. [Google Scholar] [CrossRef]
- Wang, J.; Lin, Y.H. Functionalized carbon nanotubes and nanofibers for biosensing applications. Trends Anal. Chem. 2008, 27, 619–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, I.; Heung, Y.Y.; Kim, J.H.; Lee, J.W.; Gollapudi, R.; Subramaniam, S.; Narasimhadevara, S.; Hurd, D.; Kirikera, G.R.; Shanov, V.; et al. Introduction to carbon nanotube and nanofiber smart materials. Compos. Part B Eng. 2006, 37, 382–394. [Google Scholar] [CrossRef]
- Ferancová, A.; Rassaei, L.; Marken, F.; Labuda, J.; Sillanpää, M. dsDNA modified carbon nanofiber—Solidified paste electrodes: Probing Ni(II)—dsDNA interactions. Microchim. Acta 2010, 170, 155–164. [Google Scholar] [CrossRef]
- Miller, J.N.; Miller, J.C. Statistics and Chemometrics for Analytical Chemistry, 5th ed.; Pearson Education: Essex, UK, 2005; p. 121. [Google Scholar]
- Ganguly, A.; Benson, J.; Papakonstantinou, P. Sensitive Chronocoulometric Detection of miRNA at Screen-Printed Electrodes Modified by Gold-Decorated MoS2 Nanosheets. ACS Appl. Bio Mater. 2018, 1, 1184–1194. [Google Scholar] [CrossRef]
- Martín-Yerga, D.; Costa-García, A. Towards a blocking-free electrochemical immunosensing strategy for anti-transglutaminase antibodies using screen-printed electrodes. Bioelectrochemistry 2015, 105, 88–94. [Google Scholar] [CrossRef]
- Biscay, J.; García, M.B.G.; García, A.C. Electrochemical biotin determination based on a screen printed carbon electrode array and magnetic beads. Sens. Actuators B Chem. 2014, 205, 426–432. [Google Scholar] [CrossRef]
- Erdem, A.; Congur, G.; Eksin, E. Multi channel screen printed array of electrodes for enzyme-linked voltammetric detection of microRNAs. Sens. Actuators B Chem. 2013, 188, 1089–1095. [Google Scholar] [CrossRef]
- Orum, H.; Jakobsen, M.H.; Koch, T.; Vuust, J.; Borre, M.B. Detection of the Factor V Leiden mutation by direct allele-specific hybridization of PCR amplicons to photoimmobilized locked nucleic acids. Clin. Chem. 1999, 45, 1898–1905. [Google Scholar]
- Palecek, E.; Masarik, M.; Kizek, R.; Kuhlmeier, D.; Hassmann, J.; Schülein, J. Sensitive electrochemical determination of unlabeled MutS protein and detection of point mutations in DNA. Anal. Chem. 2004, 76, 5930–5936. [Google Scholar] [CrossRef] [PubMed]
- Schimid, M.; Schalasta, G.A. A Rapid and reliable PCR based method for detecting the blood coagulation Factor V Leiden mutation. Biochemica 1997, 3, 12–15. [Google Scholar]
- Song, L.; Zhang, Y.; Li, J.; Gao, Q.; Qi, H.; Zhang, C. Non-covalent fluorescent labeling of hairpin DNA probe coupled with hybridization chain reaction for sensitive DNA detection. Appl. Spectrosc. 2016, 70, 688–694. [Google Scholar] [CrossRef]








| Ia (µA) | Ic (µA) | A (cm2) | ||
|---|---|---|---|---|
| SPE | 39.70 ± 9.02 | 29.05 ± 3.37 | 0.118 | |
| CNF-SPE | 43.8 ± 6.17 | 48.66 ± 2.37 | 0.132 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Erdem, A.; Eksin, E. Electrochemical Detection of Solution Phase Hybridization Related to Single Nucleotide Mutation by Carbon Nanofibers Enriched Electrodes. Materials 2019, 12, 3377. https://doi.org/10.3390/ma12203377
Erdem A, Eksin E. Electrochemical Detection of Solution Phase Hybridization Related to Single Nucleotide Mutation by Carbon Nanofibers Enriched Electrodes. Materials. 2019; 12(20):3377. https://doi.org/10.3390/ma12203377
Chicago/Turabian StyleErdem, Arzum, and Ece Eksin. 2019. "Electrochemical Detection of Solution Phase Hybridization Related to Single Nucleotide Mutation by Carbon Nanofibers Enriched Electrodes" Materials 12, no. 20: 3377. https://doi.org/10.3390/ma12203377