The RUBISCO to Photosystem II Ratio Limits the Maximum Photosynthetic Rate in Picocyanobacteria
Abstract
:1. Introduction
2. Experimental Section
2.1. Culturing
2.2. Whole Cell Protein Extraction and Quantitative Immunoblotting
2.3. Physiological Measurements
3. Results and Discussion
3.1. Changes in the Photosynthetic Electron Transport Chain Stoichiometry Following Subculturing

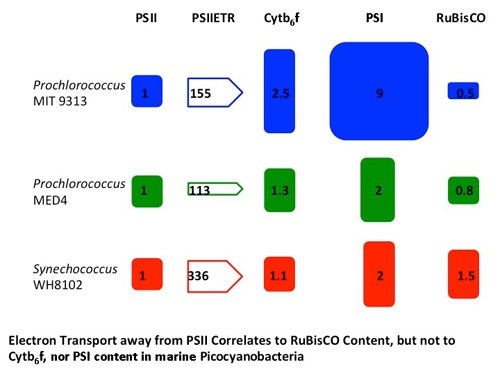
3.2. Molar Quantitations of Photosynthetic Protein Complexes

| Parameter Measured | MIT 9313 | MED 4 | WH 8102 |
|---|---|---|---|
| σPSII (Å2PSII−1) | 340 ± 17 | 312 ± 16 | 307 ± 14 |
| ETR Capacity (e-PSII−1s−1) | 155 ± 3 | 113 ± 12 | 336 ± 44 |
| ETRmax (e-PSII−1s−1) | 102 ± 2 | 98 ± 3 | 238 ± 5 |
| EK (umol photons m−2·s−1) | 54 ± 3 | 67 ± 4 | 154 ± 5 |
| μ (day−1) | 0.25 ± 0.03 | 0.19 ± 0.02 | 0.24 ± 0.02 |
| PSI:PSII | 9 ± 5 | 2 ± 0.7 | 2 ± 0.2 |
| Cytb6f:PSII | 2.5 ± 0.5 | 1.3 ± 0.4 | 1.1 ± 0.1 |
| Cytb6f:PSI | 0.2 ± 0.2 | 0.7 ± 0.1 | 0.5 ± 0.1 |
| RUBISCO:PSII | 0.5 ± 0.1 | 0.8 ± 0.2 | 1.5 ± 0.2 |
3.3. Photosystem II Photophysiological and Electron Transport Differences among Strains



3.4. Discussion
4. Conclusions
Acknowledgments
Author Contributions
Supplementary Materials
Conflicts of Interest
References
- Partensky, F.; Blanchot, J.; Vaulot, D. Differential distribution and ecology of Prochlorococcus and Synechococcus in oceanic waters: A review. Bull.-Inst. Oceanogr. Monaco-Numero Spec. 1999, 19, 457–476. [Google Scholar]
- Hess, W.R.; Rocap, G.; Ting, C.S.; Larimer, F.; Stilwagen, S.; Lamerdin, J.; Chisholm, S.W. The photosynthetic apparatus of Prochlorococcus: Insights through comparative genomics. Photosynth. Res. 2001, 70, 53–71. [Google Scholar] [CrossRef] [PubMed]
- Goericke, R.; Welschmeyer, N. The Marine Prochlorophyte Prochlorococcus Contributes Significantly to Phytoplankton Biomass and Primary Production in the Sargasso Sea. Deep Sea Res. Part I Oceanogr. Res. Pap. 1993, 40, 2283–2294. [Google Scholar] [CrossRef]
- Li, W. Composition of Ultraphytoplankton in the Central North-Atlantic. Mar. Ecol. Prog. Ser. 1995, 122, 1–8. [Google Scholar] [CrossRef]
- Liu, H.B.; Nolla, H.A.; Campbell, L. Prochlorococcus growth rate and contribution to primary production in the equatorial and subtropical North Pacific Ocean. Aquat. Microb. Ecol. 1997, 12, 39–47. [Google Scholar] [CrossRef]
- Veldhuis, M.J.W.; Kraay, G.W.; VanBleijswijk, J.D.L.; Baars, M.A. Seasonal and spatial variability in phytoplankton biomass, productivity and growth in the northwestern Indian Ocean: The southwest and northeast monsoon, 1992–1993. Deep Sea Res. Part I Oceanogr. Res. Pap. 1997, 44, 425–449. [Google Scholar] [CrossRef]
- Behrenfeld, M.J.; O’Malley, R.T.; Siegel, D.A.; McClain, C.R.; Sarmiento, J.L.; Feldman, G.C.; Milligan, A.J.; Falkowski, P.G.; Letelier, R.M.; Boss, E.S. Climate-driven trends in contemporary ocean productivity. Nature 2006, 444, 752–755. [Google Scholar] [CrossRef] [PubMed]
- Boyce, D.G.; Lewis, M.R.; Worm, B. Global phytoplankton decline over the past century. Nature 2010, 466, 591–596. [Google Scholar] [CrossRef] [PubMed]
- Irwin, A.J.; Oliver, M.J. Are ocean deserts getting larger? Geophys. Res. Lett. 2009, 36, L18609. [Google Scholar] [CrossRef]
- Ting, C.S.; Rocap, G.; King, J.; Chisholm, S.W. Cyanobacterial photosynthesis in the oceans: the origins and significance of divergent light-harvesting strategies. Trends Microbiol. 2002, 10, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Zwirglmaier, K.; Jardillier, L.; Ostrowski, M.; Mazard, S.; Garczarek, L.; Vaulot, D.; Not, F.; Massana, R.; Ulloa, O.; Scanlan, D.J. Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ. Microbiol. 2008, 10, 147–161. [Google Scholar] [PubMed]
- Rappe, M.S.; Giovannoni, S.J. The uncultured microbial majority. Annu. Rev. Microbiol. 2003, 57, 369–394. [Google Scholar] [CrossRef] [PubMed]
- Partensky, F.; Garczarek, L. Prochlorococcus: Advantages and Limits of Minimalism. Annu. Rev. Mar. Sci. 2010, 2, 305–331. [Google Scholar] [CrossRef]
- Goericke, R.; Repeta, D. Chlorophyll-a and Chlorophyll-B and Divinyl Chlorophyll-a and Chlorophyll-B in the Open Subtropical North-Atlantic Ocean. Mar. Ecol. Prog. Ser. 1993, 101, 307–313. [Google Scholar] [CrossRef]
- Morris, J.J.; Johnson, Z.I.; Szul, M.J.; Keller, M.; Zinser, E.R. Dependence of the Cyanobacterium Prochlorococcus on Hydrogen Peroxide Scavenging Microbes for Growth at the Ocean’s Surface. PLoS One 2011, 6, e16805. [Google Scholar] [CrossRef] [PubMed]
- Mella-Flores, D.; Six, C.; Ratin, M.; Partensky, F.; Boutte, C.; Le Corguille, G.; Marie, D.; Blot, N.; Gourvil, P.; Kolowrat, C.; Garczarek, L. Prochlorococcus and Synechococcus have Evolved Different Adaptive Mechanisms to Cope with Light and UV Stress. Front. Microbiol. 2012, 3. [Google Scholar] [CrossRef] [Green Version]
- Rocap, G.; Larimer, F.W.; Lamerdin, J.; Malfatti, S.; Chain, P.; Ahlgren, N.A.; Arellano, A.; Coleman, M.; Hauser, L.; Hess, W.R.; et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 2003, 424, 1042–1047. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.R.; Post, A.F.; Rocap, G.; Chisholm, S.W. Utilization of different nitrogen sources by the marine cyanobacteria Prochlorococcus and Synechococcus. Limnol. Oceanogr. 2002, 47, 989–996. [Google Scholar] [CrossRef]
- Dufresne, A.; Garczarek, L.; Partensky, F. Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol. 2005, 6, R14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kettler, G.C.; Martiny, A.C.; Huang, K.; Zucker, J.; Coleman, M.L.; Rodrigue, S.; Chen, F.; Lapidus, A.; Ferriera, S.; Johnson, J.; et al. Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. Plos Genet. 2007, 3, 2515–2528. [Google Scholar] [CrossRef]
- Zhaxybayeva, O.; Doolittle, W.F.; Papke, R.T.; Gogarten, J.P. Intertwined Evolutionary Histories of Marine Synechococcus and Prochlorococcus marinus. Genome Biol. Evol. 2009, 1, 325–339. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Falkowski, P.G. Genome evolution in cyanobacteria: The stable core and the variable shell. Proc. Natl. Acad. Sci. USA 2008, 105, 2510–2515. [Google Scholar] [CrossRef] [PubMed]
- Bibby, T.S.; Mary, I.; Nield, J.; Partensky, F.; Barber, J. Low-light-adapted Prochlorococcus species possess specific antennae for each photosystem. Nature 2003, 424, 1051–1054. [Google Scholar] [CrossRef] [PubMed]
- Tolonen, A.C.; Aach, J.; Lindell, D.; Johnson, Z.I.; Rector, T.; Steen, R.; Church, G.M.; Chisholm, S.W. Global gene expression of Prochlorococcus ecotypes in response to changes in nitrogen availability. Mol. Syst. Biol. 2006, 2. [Google Scholar] [CrossRef]
- Waldbauer, J.R.; Rodrigue, S.; Coleman, M.L.; Chisholm, S.W. Transcriptome and Proteome Dynamics of a Light-Dark Synchronized Bacterial Cell Cycle. PLoS One 2012, 7, e43432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campbell, D.A.; Cockshutt, A.M.; Porankiewicz-Asplund, J. Analysing photosynthetic complexes in uncharacterized species or mixed microalgal communities using global antibodies. Physiol. Plant. 2003, 119, 322–327. [Google Scholar] [CrossRef]
- Gygi, S.P.; Rochon, Y.; Franza, B.R.; Aebersold, R. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 1999, 19, 1720–1730. [Google Scholar] [PubMed]
- Maier, T.; Schmidt, A.; Güell, M.; Kühner, S.; Gavin, A.-C.; Aebersold, R.; Serrano, L. Quantification of mRNA and protein and integration with protein turnover in a bacterium. Mol. Syst. Biol. 2011, 7. [Google Scholar] [CrossRef]
- Six, C.; Finkel, Z.V.; Irwin, A.J.; Campbell, D.A. Light Variability Illuminates Niche-Partitioning among Marine Picocyanobacteria. PloS One 2007, 2, e1341. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.M.; MacKinnon, J.D.; Cockshutt, A.M.; Villareal, T.A.; Campbell, D.A. Flux capacities and acclimation costs in Trichodesmium from the Gulf of Mexico. Mar. Biol. 2008, 154, 413–422. [Google Scholar] [CrossRef]
- Kolber, Z.S.; Prášil, O.; Falkowski, P.G. Measurements of variable chlorophyll fluorescence using fast repetition rate techniques: Defining methodology and experimental protocols. Biochim. Biophys. Acta—Bioenerg. 1998, 1367, 88–106. [Google Scholar] [CrossRef]
- Laney, S.R.; Letelier, R.M. Artifacts in measurements of chlorophyll fluorescence transients, with specific application to fast repetition rate fluorometry. Limnol. Oceanogr. Methods 2008, 6, 40–50. [Google Scholar] [CrossRef]
- Suggett, D.; MacIntyre, H.; Kana, T.; Geider, R. Comparing electron transport with gas exchange: Parameterising exchange rates between alternative photosynthetic currencies for eukaryotic phytoplankton. Aquat. Microb. Ecol. 2009, 56, 147–162. [Google Scholar] [CrossRef]
- Jassby, A.D.; Platt, T. Mathematical formulation of the relationship between photosynthesis and light for phytoplankton. Limnol. Oceanogr. 1976, 21, 540–548. [Google Scholar] [CrossRef]
- Laisk, A.; Eichelmann, H.; Oja, V.; Peterson, R.B. Control of cytochrome b6f at low and high light intensity and cyclic electron transport in leaves. Biochim. Biophys. Acta—Bioenerg. 2005, 1708, 79–90. [Google Scholar] [CrossRef]
- Tikkanen, M.; Grieco, M.; Nurmi, M.; Rantala, M.; Suorsa, M.; Aro, E.-M. Regulation of the photosynthetic apparatus under fluctuating growth light. Philos. Trans. R. Soc. B-Biol. Sci. 2012, 367, 3486–3493. [Google Scholar]
- Fraser, J.M.; Tulk, S.E.; Jeans, J.A.; Campbell, D.A.; Bibby, T.S.; Cockshutt, A.M. Photophysiological and Photosynthetic Complex Changes during Iron Starvation in Synechocystis sp. PCC 6803 and Synechococcus elongatus PCC 7942. Plos One 2013, 8, e59861. [Google Scholar] [CrossRef] [PubMed]
- Sukenik, A.; Bennett, J.; Falkowski, P. Light-saturated photosynthesis—Limitation by electron transport or carbon fixation? Biochim. Biophys. Acta—Bioenerg. 1987, 891, 205–215. [Google Scholar] [CrossRef]
- Vandenhecke, J.M.; Bastedo, J.; Cockshutt, A.M.; Campbell, D.A.; Huot, Y. Changes in the Rubisco to Photosystem ratio dominate photoacclimation across phytoplankton taxa. Photosythesis Res. 2015. Submitted for publication. [Google Scholar]
- Kulk, G.; de Vries, P.; van de Poll, W.H.; Visser, R.J.W.; Buma, A.G.J. Temperature-dependent growth and photophysiology of prokaryotic and eukaryotic oceanic picophytoplankton. Mar. Ecol. Prog. Ser. 2012, 466, 43–55. [Google Scholar] [CrossRef]
- Umena, Y.; Kawakami, K.; Shen, J.-R.; Kamiya, N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 2011, 473, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Brunger, A.T.; Adams, P.D.; Fromme, P.; Fromme, R.; Levitt, M.; Schröder, G.F. Improving the accuracy of macromolecular structure refinement at 7 Å resolution. Structure 2012, 20, 957–966. [Google Scholar] [CrossRef] [PubMed]
- Kurisu, G.; Zhang, H.; Smith, J.L.; Cramer, W.A. Structure of the cytochrome b6f complex of oxygenic photosynthesis: tuning the cavity. Science 2003, 302, 1009–1014. [Google Scholar] [CrossRef] [PubMed]
- Behrenfeld, M.J.; Milligan, A.J. Photophysiological Expressions of Iron Stress in Phytoplankton. Annu. Rev. Mar. Sci. 2013, 5, 217–246. [Google Scholar] [CrossRef]
- Rusch, D.B.; Martiny, A.C.; Dupont, C.L.; Halpern, A.L.; Venter, J.C. Characterization of Prochlorococcus clades from iron-depleted oceanic regions. Proc. Natl. Acad. Sci. USA 2010, 107, 16184–16189. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zorz, J.K.; Allanach, J.R.; Murphy, C.D.; Roodvoets, M.S.; Campbell, D.A.; Cockshutt, A.M. The RUBISCO to Photosystem II Ratio Limits the Maximum Photosynthetic Rate in Picocyanobacteria. Life 2015, 5, 403-417. https://doi.org/10.3390/life5010403
Zorz JK, Allanach JR, Murphy CD, Roodvoets MS, Campbell DA, Cockshutt AM. The RUBISCO to Photosystem II Ratio Limits the Maximum Photosynthetic Rate in Picocyanobacteria. Life. 2015; 5(1):403-417. https://doi.org/10.3390/life5010403
Chicago/Turabian StyleZorz, Jackie K., Jessica R. Allanach, Cole D. Murphy, Mitchell S. Roodvoets, Douglas A. Campbell, and Amanda M. Cockshutt. 2015. "The RUBISCO to Photosystem II Ratio Limits the Maximum Photosynthetic Rate in Picocyanobacteria" Life 5, no. 1: 403-417. https://doi.org/10.3390/life5010403