Functionalized Nanocellulose Drives Neural Stem Cells toward Neuronal Differentiation
Abstract
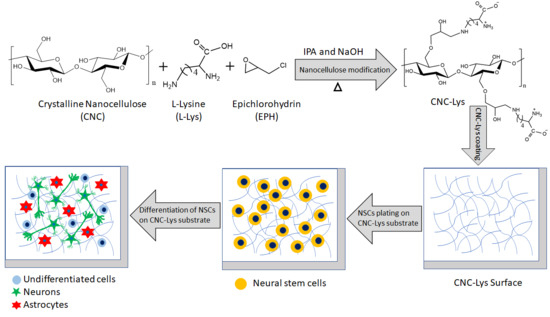
:1. Introduction
2. Results
2.1. Characterization of the CNC–Lys Material
2.2. Proliferation and Differentiation of NSCs/NPCs

2.3. The Majority of the rNPCs Differentiated into Neurons on the CNC–Lys Surface

2.4. Electrophysiological Activities of Neurons Differentiated on CNC–Lys
3. Discussion
4. Materials and Methods
4.1. Reagents and Chemicals
4.2. Preparation of the Nanocellulose Surface (CNC–Lys)

4.3. Characterization of CNC–Lys
4.4. Culture of Rat Neural Stem Cells (rNSCs) and Their Propagation and Passage
4.5. Preparation of Culture Surfaces
4.6. Differentiation of Neural Progenitor Cells (NPCs)
4.7. Scanning Electron Microscopy (SEM) for Morphological Examination of Differentiated rNSCs on the CNC–Lys Surface
4.8. Immunostaining and Imaging
4.9. Plating of Cells in CNC-, PDL-, and CNC–Lys-Coated MEA Dishes
4.10. Recording from the MEA Dishes
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jellinger, K.A.; Stadelmann, C. The enigma of cell death in neurodegenerative disorders. J. Neural Transm. Suppl. 2000, 60, 21–36. [Google Scholar]
- Gorman, A.M. Neuronal cell death in neurodegenerative diseases: Recurring themes around protein handling. J. Cell. Mol. Med. 2008, 12, 2263–2280. [Google Scholar] [CrossRef]
- Ludwig, P.E.; Thankam, F.G.; Patil, A.A.; Chamczuk, A.J.; Agrawal, D.K. Brain injury and neural stem cells. Neural Regen. Res. 2018, 13, 7–18. [Google Scholar]
- Weston, N.M.; Sun, D. The potential of stem cells in treatment of traumatic brain injury. Curr. Neurol. Neurosci. Rep. 2018, 18, 1. [Google Scholar] [CrossRef] [PubMed]
- Raisman, G. Neural stem cells for brain and spinal cord repair. J. Neurol. Neurosurg. Psychiatry 2003, 74, 1602. [Google Scholar] [CrossRef] [Green Version]
- Miller, R.H. The promise of stem cells for neural repair. Brain Res. 2006, 1091, 258–264. [Google Scholar] [CrossRef]
- Webber, D.J.; Minger, S.L. Therapeutic potential of stem cells in central nervous system regeneration. Curr. Opin. Investig. Drugs 2004, 5, 714–719. [Google Scholar]
- Chang, Y.L.; Chen, S.J.; Kao, C.L.; Hung, S.C.; Ding, D.C.; Yu, C.C.; Chen, Y.J.; Ku, H.H.; Lin, C.P.; Lee, K.H.; et al. Docosahexaenoic acid promotes dopaminergic differentiation in induced pluripotent stem cells and inhibits teratoma formation in rats with Parkinson-like pathology. Cell Transplant. 2012, 21, 313–332. [Google Scholar] [CrossRef]
- Steward, O.; Sharp, K.G.; Yee, K.M.; Hatch, M.N.; Bonner, J.F. Characterization of ectopic colonies that form in widespread areas of the nervous system with neural stem cell transplants into the site of a severe spinal cord injury. J. Neurosci. 2014, 34, 14013–14021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, L.H.; Björklund, A. Reconstruction of brain circuitry by neural transplants generated from pluripotent stem cells. Neurobiol. Dis. 2015, 79, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Long, K.R.; Huttner, W.B. How the extracellular matrix shapes neural development. Open Biol. 2019, 9, 180216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hynes, R.O. The extracellular matrix: Not just pretty fibrils. Science 2009, 326, 1216–1219. [Google Scholar] [CrossRef] [Green Version]
- Navarro Quiroz, E.; Navarro Quiroz, R.; Ahmad, M.; Gomez Escorcia, L.; Villarreal, J.; Fernandez Ponce, C.; Aroca Martinez, G. Cell signaling in neuronal stem cells. Cells 2018, 7, 75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vieira, M.S.; Goulart, V.A.M.; Parreira, R.C.; Oliveira-Lima, O.C.; Glaser, T.; Naaldijk, Y.M.; Ferrer, A.; Savanur, V.H.; Reyes, P.A.; Sandiford, O.; et al. Decoding epigenetic cell signaling in neuronal differentiation. Semin. Cell Dev. Biol. 2019, 95, 12–24. [Google Scholar] [CrossRef] [PubMed]
- De, M.; Ghosh, P.S.; Rotello, V.M. Applications of nanoparticles in biology. Adv. Mater. 2008, 20, 4225–4241. [Google Scholar] [CrossRef] [Green Version]
- Gil, P.R.; Yang, F.; Thomas, H.; Li, L.; Terfort, A.; Parak, W.J. Development of an assay based on cell counting with quantum dot labels for comparing cell adhesion within cocultures. Nano Today 2011, 6, 20–27. [Google Scholar]
- Rivera Gil, P.; Hühn, D.; del Mercato, L.L.; Sasse, D.; Parak, W.J. Nanopharmacy: Inorganic nanoscale devices as vectors and active compounds. Pharmacol. Res. 2010, 62, 115–125. [Google Scholar] [CrossRef]
- Tong, X.; Yang, F. Engineering interpenetrating network hydrogels as biomimetic cell niche with independently tunable biochemical and mechanical properties. Biomaterials 2014, 35, 1807–1815. [Google Scholar] [CrossRef]
- Khan, F.; Tanaka, M. Designing smart biomaterials for tissue engineering. Int. J. Mol. Sci. 2018, 19, 17. [Google Scholar] [CrossRef] [Green Version]
- Beachley, V.; Wen, X. Polymer nanofibrous structures: Fabrication, biofunctionalization, and cell interactions. Prog. Polym. Sci. 2010, 35, 868–892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kin, S.; Hagiwara, A.; Nakase, Y.; Kuriu, Y.; Nakashima, S.; Yoshikawa, T.; Sakakura, C.; Otsuji, E.; Nakamura, T.; Yamagishi, H. Regeneration of skeletal muscle using in situ tissue engineering on an acellular collagen sponge scaffold in a rabbit model. ASAIO J. 2007, 53, 506–513. [Google Scholar] [CrossRef] [PubMed]
- Klumpp, D.; Horch, R.E.; Kneser, U.; Beier, J.P. Engineering skeletal muscle tissue—New perspectives in vitro and in vivo. J. Cell. Mol. Med. 2010, 14, 2622–2629. [Google Scholar] [CrossRef] [PubMed]
- Sicari, B.M.; Dearth, C.L.; Badylak, S.F. Tissue engineering and regenerative medicine approaches to enhance the functional response to skeletal muscle injury. Anat. Rec. 2014, 297, 51–64. [Google Scholar] [CrossRef]
- Pina, S.; Oliveira, J.M.; Reis, R.L. Natural-based nanocomposites for bone tissue engineering and regenerative medicine: A review. Adv. Mater. 2015, 27, 1143–1169. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Krishnan, U.M.; Sethuraman, S. Development of biomaterial scaffold for nerve tissue engineering: Biomaterial mediated neural regeneration. J. Biomed. Sci. 2009, 16, 108. [Google Scholar] [CrossRef] [Green Version]
- Klemm, D.; Heublein, B.; Fink, H.P.; Bohn, A. Cellulose: Fascinating biopolymer and sustainable raw material. Angew. Chem. Int. Ed. 2005, 44, 3358–3393. [Google Scholar] [CrossRef] [PubMed]
- Endes, C.; Camarero-Espinosa, S.; Mueller, S.; Foster, E.J.; Petri-Fink, A.; Rothen-Rutishauser, B.; Weder, C.; Clift, M.J.D. A critical review of the current knowledge regarding the biological impact of nanocellulose. J. Nanobiotechnol. 2016, 14, 78. [Google Scholar] [CrossRef] [Green Version]
- Chhetri, B.P.; Soni, D.; Rangumagar, A.B.; Parnell, C.M.; Wayland, H.; Watanabe, F.; Kannarpady, G.; Biris, A.S.; Ghosh, A. Synthesis, characterization, and photocatalytic activity of N-doped carbonaceous material derived from cellulose in textile dye remediation. J. Environ. Chem. Eng. 2017, 5, 2586–2596. [Google Scholar] [CrossRef]
- Picheth, G.F.; Pirich, C.L.; Sierakowski, M.R.; Woehl, M.A.; Sakakibara, C.N.; de Souza, C.F.; Martin, A.A.; da Silva, R.; de Freitas, R.A. Bacterial cellulose in biomedical applications: A review. Int. J. Biol. Macromol. 2017, 104, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Tu, C.-W.; Tsai, F.-C.; Chen, J.-K.; Wang, H.-P.; Lee, R.-H.; Zhang, J.; Chen, T.; Wang, C.-C.; Huang, C.-F. Preparations of Tough and Conductive PAMPS/PAA Double Network Hydrogels Containing Cellulose Nanofibers and Polypyrroles. Polymers 2020, 12, 2835. [Google Scholar] [CrossRef]
- Tu, C.-W.; Tsai, F.-C.; Chang, C.-J.; Yang, C.-H.; Kuo, S.-W.; Zhang, J.; Chen, T.; Huang, C.-F. Surface-initiated initiators for continuous activator regeneration (SI ICAR) ATRP of MMA from 2,2,6,6–tetramethylpiperidine–1–oxy (TEMPO) oxidized cellulose nanofibers for the preparations of PMMA nanocomposites. Polymers 2019, 11, 1631. [Google Scholar] [CrossRef] [Green Version]
- Klemm, D.; Kramer, F.; Moritz, S.; Lindström, T.; Ankerfors, M.; Gray, D.; Dorris, A. Nanocelluloses: A new family of nature-based materials. Angew. Chem. Int. Ed. 2011, 50, 5438–5466. [Google Scholar] [CrossRef]
- Dufresne, A. Nanocellulose: From Nature to High Performance Tailored Materials; De Gruyter Mouton: Berlin, Germany, 2012; Volume 67, ISBN 9783110254600. [Google Scholar]
- Lin, N.; Dufresne, A. Nanocellulose in biomedicine: Current status and future prospect. Eur. Polym. J. 2014, 59, 302–325. [Google Scholar] [CrossRef] [Green Version]
- Jorfi, M.; Foster, E.J. Recent advances in nanocellulose for biomedical applications. J. Appl. Polym. Sci. 2015, 132, 41719. [Google Scholar] [CrossRef]
- Norman, J.J.; Desai, T.A. Methods for fabrication of nanoscale topography for tissue engineering scaffolds. Ann. Biomed. Eng. 2006, 34, 89–101. [Google Scholar] [CrossRef] [PubMed]
- McCracken, K.E.; Tran, P.L.; You, D.J.; Slepian, M.J.; Yoon, J.Y. Shear- vs. nanotopography-guided control of growth of endothelial cells on RGD-nanoparticle-nanowell arrays. J. Biol. Eng. 2013, 7, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, Y.; Gardner, D.J.; Han, Y.; Kiziltas, A.; Cai, Z.; Tshabalala, M.A. Influence of drying method on the material properties of nanocellulose I: Thermostability and crystallinity. Cellulose 2013, 20, 2379–2392. [Google Scholar] [CrossRef]
- Shelke, N.B.; James, R.; Laurencin, C.T.; Kumbar, S.G. Polysaccharide biomaterials for drug delivery and regenerative engineering. Polym. Adv. Technol. 2014, 25, 448–460. [Google Scholar] [CrossRef]
- Dadsetan, M.; Pumberger, M.; Casper, M.E.; Shogren, K.; Giuliani, M.; Ruesink, T.; Hefferan, T.E.; Currier, B.L.; Yaszemski, M.J. The effects of fixed electrical charge on chondrocyte behavior. Acta Biomater. 2011, 7, 2080–2090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, H.-I.; Wang, Y. Cell responses to surface and architecture of tissue engineering scaffolds. In Regenerative Medicine and Tissue Engineering—Cells and Biomaterials; InTech: London, UK, 2011. [Google Scholar]
- Sergeeva, Y.N.; Huang, T.; Felix, O.; Jung, L.; Tropel, P.; Viville, S.; Decher, G. What is really driving cell–surface interactions? Layer-by-layer assembled films may help to answer questions concerning cell attachment and response to biomaterials. Biointerphases 2016, 11, 019009. [Google Scholar] [CrossRef]
- Bhattacharya, M.; Malinen, M.M.; Lauren, P.; Lou, Y.R.; Kuisma, S.W.; Kanninen, L.; Lille, M.; Corlu, A.; Guguen-Guillouzo, C.; Ikkala, O.; et al. Nanofibrillar cellulose hydrogel promotes three-dimensional liver cell culture. J. Control. Release 2012, 164, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Zander, N.E.; Dong, H.; Steele, J.; Grant, J.T. Metal cation cross-linked nanocellulose hydrogels as tissue engineering substrates. ACS Appl. Mater. Interfaces 2014, 6, 18502–18510. [Google Scholar] [CrossRef] [PubMed]
- Evans, N.; Minelli, C.; Gentleman, E.; LaPointe, V.; Patankar, S.; Kallivretaki, M.; Chen, X.; Roberts, C.; Stevens, M. Substrate stiffness affects early differentiation events in embryonic stem cells. Eur. Cells Mater. 2009, 18, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Pelton, R. Bioactive paper provides a low-cost platform for diagnostics. TrAC—Trends Anal. Chem. 2009, 28, 925–942. [Google Scholar] [CrossRef]
- Dong, S.; Roman, M. Fluorescently labeled cellulose nanocrystals for bioimaging applications. J. Am. Chem. Soc. 2007, 129, 13810–13811. [Google Scholar] [CrossRef]
- Chai, C.; Leong, K.W. Biomaterials approach to expand and direct differentiation of stem cells. Mol. Ther. 2007, 15, 467–480. [Google Scholar] [CrossRef]
- Claverol-Tinturé, E.; Cabestany, J.; Rosell, X. Multisite recording of extracellular potentials produced by microchannel-confined neurons in-vitro. IEEE Trans. Biomed. Eng. 2007, 54, 331–335. [Google Scholar] [CrossRef] [Green Version]
- Claverol-Tinturé, E.; Ghirardi, M.; Fiumara, F.; Rosell, X.; Cabestany, J. Multielectrode arrays with elastomeric microstructured overlays for extracellular recordings from patterned neurons. J. Neural Eng. 2005, 2, L1. [Google Scholar] [CrossRef]
- Morin, F.O.; Takamura, Y.; Tamiya, E. Investigating neuronal activity with planar microelectrode arrays: Achievements and new perspectives. J. Biosci. Bioeng. 2005, 100, 131–143. [Google Scholar] [CrossRef]
- Ito, D.; Tamate, H.; Nagayama, M.; Uchida, T.; Kudoh, S.N.; Gohara, K. Minimum neuron density for synchronized bursts in a rat cortical culture on multi-electrode arrays. Neuroscience 2010, 171, 50–61. [Google Scholar] [CrossRef]
- Plenz, D.; Stewart, C.V.; Shew, W.; Yang, H.; Klaus, A.; Bellay, T. Multi-electrode array recordings of neuronal avalanches in organotypic cultures. J. Vis. Exp. 2011, 54, e2949. [Google Scholar] [CrossRef]
- Bardy, C.; Van Den Hurk, M.; Eames, T.; Marchand, C.; Hernandez, R.V.; Kellogg, M.; Gorris, M.; Galet, B.; Palomares, V.; Brown, J.; et al. Neuronal medium that supports basic synaptic functions and activity of human neurons in vitro. Proc. Natl. Acad. Sci. USA 2015, 112, E2725–E2734. [Google Scholar] [CrossRef] [Green Version]
- Saalfrank, D.; Konduri, A.K.; Latifi, S.; Habibey, R.; Golabchi, A.; Martiniuc, A.V.; Knoll, A.; Ingebrandt, S.; Blau, A. Incubator-independent cell-culture perfusion platform for continuous long-term microelectrode array electrophysiology and time-lapse imaging. R. Soc. Open Sci. 2015, 2, 150031. [Google Scholar] [CrossRef]
- Amin, H.; Maccione, A.; Marinaro, F.; Zordan, S.; Nieus, T.; Berdondini, L. Electrical responses and spontaneous activity of human iPS-derived neuronal networks characterized for 3-month culture with 4096-electrode arrays. Front. Neurosci. 2016, 10, 212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seidel, D.; Jahnke, H.G.; Englich, B.; Girard, M.; Robitzki, A.A. In vitro field potential monitoring on a multi-microelectrode array for the electrophysiological long-term screening of neural stem cell maturation. Analyst 2017, 142, 1929–1937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pine, J. Recording action potentials from cultured neurons with extracellular microcircuit electrodes. J. Neurosci. Methods 1980, 2, 19–31. [Google Scholar] [CrossRef]
- Taketani, M.; Baudry, M. Advances in Network Electrophysiology: Using Multi-Electrode Arrays; Springer: Boston, MA, USA, 2006; ISBN 9780387258584. [Google Scholar]
- Park, S.; Baker, J.O.; Himmel, M.E.; Parilla, P.A.; Johnson, D.K. Cellulose crystallinity index: Measurement techniques and their impact on interpreting cellulase performance. Biotechnol. Biofuels 2010, 3, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Garvey, C.J.; Parker, I.H.; Simon, G.P. On the interpretation of X-ray diffraction powder patterns in terms of the nanostructure of cellulose I fibres. Macromol. Chem. Phys. 2005, 206, 1568–1575. [Google Scholar] [CrossRef]
- He, J.; Cui, S.; Wang, S.Y. Preparation and crystalline analysis of high-grade bamboo dissolving pulp for cellulose acetate. J. Appl. Polym. Sci. 2008, 107, 1029–1038. [Google Scholar] [CrossRef]
- Ross, A.M.; Jiang, Z.; Bastmeyer, M.; Lahann, J. Physical aspects of cell culture substrates: Topography, roughness, and elasticity. Small 2012, 8, 336–355. [Google Scholar] [CrossRef] [Green Version]
- Anirudhan, T.S.; Rejeena, S.R. Adsorption and hydrolytic activity of trypsin on a carboxylate-functionalized cation exchanger prepared from nanocellulose. J. Colloid Interface Sci. 2012, 381, 125–136. [Google Scholar] [CrossRef]
- Kong, J.; Yu, S. Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta Biochim. Biophys. Sin. 2007, 39, 549–559. [Google Scholar] [CrossRef] [Green Version]
- Nyström, G.; Mihranyan, A.; Razaq, A.; Lindström, T.; Nyholm, L.; Strømme, M. A nanocellulose polypyrrole composite based on microfibrillated cellulose from wood. J. Phys. Chem. B 2010, 114, 4178–4182. [Google Scholar] [CrossRef]
- Aydin, M.; Kartal, Z.; Osmanoǧlu, Ş.; Halim Başkan, M.; Topkaya, R. EPR and FT–IR spectroscopic studies of l-lysine monohydrochloride and l-glutamic acid hydrochloride powders. J. Mol. Struct. 2011, 994, 150–154. [Google Scholar] [CrossRef]
- Courtenay, J.C.; Johns, M.A.; Galembeck, F.; Deneke, C.; Lanzoni, E.M.; Costa, C.A.; Scott, J.L.; Sharma, R.I. Surface modified cellulose scaffolds for tissue engineering. Cellulose 2017, 24, 253–267. [Google Scholar] [CrossRef] [Green Version]
- Moon, R.J.; Martini, A.; Nairn, J.; Simonsen, J.; Youngblood, J. Cellulose nanomaterials review: Structure, properties and nanocomposites. Chem. Soc. Rev. 2011, 40, 3941–3994. [Google Scholar] [CrossRef] [PubMed]
- Saha, K.; Keung, A.J.; Irwin, E.F.; Li, Y.; Little, L.; Schaffer, D.V.; Healy, K.E. Substrate modulus directs neural stem cell behavior. Biophys. J. 2008, 95, 4426–4438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, H.; Luo, Q.; Zhou, D. Affinity membrane chromatography for the analysis and purification of proteins. J. Biochem. Biophys. Methods 2001, 49, 199–240. [Google Scholar] [CrossRef]
- Wu, C.Y.; Suen, S.Y.; Chen, S.C.; Tzeng, J.H. Analysis of protein adsorption on regenerated cellulose-based immobilized copper ion affinity membranes. J. Chromatogr. A 2003, 996, 53–70. [Google Scholar] [CrossRef]
- Innala, M.; Riebe, I.; Kuzmenko, V.; Sundberg, J.; Gatenholm, P.; Hanse, E.; Johannesson, S. 3D Culturing and differentiation of SH-SY5Y neuroblastoma cells on bacterial nanocellulose scaffolds. Artif. Cells Nanomed. Biotechnol. 2014, 42, 302–308. [Google Scholar] [CrossRef]
- Jonsson, M.; Brackmann, C.; Puchades, M.; Brattås, K.; Ewing, A.; Gatenholm, P.; Enejder, A. Neuronal Networks on Nanocellulose Scaffolds. Tissue Eng.—Part C Methods 2015, 21, 1162–1170. [Google Scholar] [CrossRef]
- Stabenfeldt, S.E.; Munglani, G.; García, A.J.; Laplaca, M.C. Biomimetic microenvironment modulates neural stem cell survival, migration, and differentiation. Tissue Eng.—Part A 2010, 16, 3747–3758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schubert, D.; Humphreys, S.; de Vitry, F.; Jacob, F. Induced differentiation of a neuroblastoma. Dev. Biol. 1971, 25, 514–546. [Google Scholar] [CrossRef]
- Min, S.K.; Jung, S.M.; Ju, J.H.; Kwon, Y.S.; Yoon, G.H.; Shin, H.S. Regulation of astrocyte activity via control over stiffness of cellulose acetate electrospun nanofiber. Vitr. Cell. Dev. Biol.—Anim. 2015, 51, 933–940. [Google Scholar] [CrossRef]
- Pankevich, D.E.; Davis, M.; Altevogt, B.M. Glutamate-Related Biomarkers in Drug Development for Disorders of the Nervous System; National Academies Press: Washington, DC, USA, 2011. [Google Scholar]
- Herzog, E.; Bellenchi, G.C.; Gras, C.; Bernard, V.; Ravassard, P.; Bedet, C.; Gasnier, B.; Giros, B.; El Mestikawy, S. The existence of a second vesicular glutamate transporter specifies subpopulations of glutamatergic neurons. J. Neurosci. 2001, 21, RC181. [Google Scholar] [CrossRef] [PubMed]
- Bernard, C.; Cossart, R.; Hirsch, J.C.; Esclapez, M.; Ben-Ari, Y. What is GABAergic Inhibition? How Is it Modified in Epilepsy? Epilepsia 2000, 41, S90–S95. [Google Scholar] [CrossRef]
- Hindley, C.; Ali, F.; McDowell, G.; Cheng, K.; Jones, A.; Guillemot, F.; Philpott, A. Post-translational modification of Ngn2 differentially affects transcription of distinct targets to regulate the balance between progenitor maintenance and differentiation. Development 2012, 139, 1718–1723. [Google Scholar] [CrossRef] [Green Version]
- Corner, M.A.; Van Pelt, J.; Wolters, P.S.; Baker, R.E.; Nuytinck, R.H. Physiological effects of sustained blockade of excitatory synaptic transmission on spontaneously active developing neuronal networks—An inquiry into the reciprocal linkage between intrinsic biorhythms and neuroplasticity in early ontogeny. Neurosci. Biobehav. Rev. 2002, 26, 127–185. [Google Scholar] [CrossRef]
- Van Pelt, J.; Wolters, P.S.; Corner, M.A.; Rutten, W.L.C.; Ramakers, G.J.A. Long-term characterization of firing dynamics of spontaneous bursts in cultured neural networks. IEEE Trans. Biomed. Eng. 2004, 51, 2051–2062. [Google Scholar] [CrossRef] [PubMed]
- Demas, J.; Eglen, S.J.; Wong, R.O.L. Developmental loss of synchronous spontaneous activity in the mouse retina is independent of visual experience. J. Neurosci. 2003, 23, 2851–2860. [Google Scholar] [CrossRef] [Green Version]
- Lestienne, R. Spike timing, synchronization and information processing on the sensory side of the central nervous system. Prog. Neurobiol. 2001, 65, 545–591. [Google Scholar] [CrossRef]
- Limpert, E.; Stahel, W.A.; Abbt, M. Log-normal distributions across the sciences: Keys and clues on the charms of statistics, and how mechanical models resembling gambling machines offer a link to a handy way to characterize log-normal distributions, which can provide deeper insight into var. Bioscience 2001, 51, 341–352. [Google Scholar] [CrossRef]
- Van Pelt, J.; Vajda, I.; Wolters, P.S.; Corner, M.A.; Ramakers, G.J.A. Dynamics and plasticity in developing neuronal networks in vitro. Prog. Brain Res. 2005, 147, 171–188. [Google Scholar]
- Chiappalone, M.; Bove, M.; Vato, A.; Tedesco, M.; Martinoia, S. Dissociated cortical networks show spontaneously correlated activity patterns during in vitro development. Brain Res. 2006, 1093, 41–53. [Google Scholar] [CrossRef]
- Chiappalone, M.; Vato, A.; Berdondini, L.; Koudelka-Hep, M.; Martinoia, S. Network dynamics and synchronous activity in cultured cortical neurons. Int. J. Neural Syst. 2007, 17, 87–103. [Google Scholar] [CrossRef]
- Czarnecki, A.; Tscherter, A.; Streit, J. Network activity and spike discharge oscillations in cortical slice cultures from neonatal rat. Eur. J. Neurosci. 2012, 35, 375–388. [Google Scholar] [CrossRef] [PubMed]
- Ban, J.; Bonifazi, P.; Pinato, G.; Broccard, F.; Studer, L.; Torre, V.; Ruaro, M.E. ES-derived neurons form functional networks in vitro. Stem Cells 2006, 25, 738–749. [Google Scholar] [CrossRef] [PubMed]
- Beske, P.H.; Scheeler, S.M.; Adler, M.; McNutt, P.M. Accelerated intoxication of GABAergic synapses by botulinum neurotoxin A disinhibits stem cell-derived neuron networks prior to network silencing. Front. Cell. Neurosci. 2015, 9, 159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, D.; Komatsu, T.; Gohara, K. Measurement of saturation processes in glutamatergic and GABAergic synapse densities during long-term development of cultured rat cortical networks. Brain Res. 2013, 1534, 22–32. [Google Scholar] [CrossRef]
- Muramoto, K.; Ichikawa, M.; Kawahara, M.; Kobayashi, K.; Kuroda, Y. Frequency of synchronous oscillations of neuronal activity increases during development and is correlated to the number of synapses in cultured cortical neuron networks. Neurosci. Lett. 1993, 163, 163–165. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, X.; Li, B. Adsorption of Hg2+ and Cd2+ by ethylenediamine modified peanut shells. Carbohydr. Polym. 2010, 81, 335–339. [Google Scholar] [CrossRef]
- Ding, Z.; Yu, R.; Hu, X.; Chen, Y.; Zhang, Y. Graft copolymerization of epichlorohydrin and ethylenediamine onto cellulose derived from agricultural by-products for adsorption of Pb(II) in aqueous solution. Cellulose 2014, 21, 1459–1469. [Google Scholar] [CrossRef]
- Udoetok, I.A.; Dimmick, R.M.; Wilson, L.D.; Headley, J.V. Adsorption properties of cross-linked cellulose-epichlorohydrin polymers in aqueous solution. Carbohydr. Polym. 2016, 136, 329–340. [Google Scholar] [CrossRef]
- Bonaguidi, M.A.; Peng, C.Y.; McGuire, T.; Falciglia, G.; Gobeske, K.T.; Czeisler, C.; Kessler, J.A. Noggin expands neural stem cells in the adult hippocampus. J. Neurosci. 2008, 28, 9194–9204. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Huang, Z.; Liu, W.; Yang, K.; Sun, K.; Xing, S.; Wang, D.; Zhang, W.; Jiang, X. Surface coating as a key parameter in engineering neuronal network structures in vitro. Biointerphases 2012, 7, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Potter, S.M.; DeMarse, T.B. A new approach to neural cell culture for long-term studies. J. Neurosci. Methods 2001, 110, 17–24. [Google Scholar] [CrossRef]





| Culture Substratum | Total Cells Plated/Dish | Cell Counts | ||
|---|---|---|---|---|
| Week 1 | Week 2 | Week 3 | ||
| CNC | 10,000 | 3450 ± 160.33 | 3374 ± 137.72 | 2561 ± 99.79 |
| PDL | 10,000 | 8531 ± 149.81 | 8188 ± 118.15 | 8150 ± 120.56 |
| CNC–Lys | 10,000 | 9423 ± 109.4 | 8329 ± 251.1 | 8199 ± 177.06 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pandanaboina, S.C.; RanguMagar, A.B.; Sharma, K.D.; Chhetri, B.P.; Parnell, C.M.; Xie, J.Y.; Srivatsan, M.; Ghosh, A. Functionalized Nanocellulose Drives Neural Stem Cells toward Neuronal Differentiation. J. Funct. Biomater. 2021, 12, 64. https://doi.org/10.3390/jfb12040064
Pandanaboina SC, RanguMagar AB, Sharma KD, Chhetri BP, Parnell CM, Xie JY, Srivatsan M, Ghosh A. Functionalized Nanocellulose Drives Neural Stem Cells toward Neuronal Differentiation. Journal of Functional Biomaterials. 2021; 12(4):64. https://doi.org/10.3390/jfb12040064
Chicago/Turabian StylePandanaboina, Sahitya Chetan, Ambar B. RanguMagar, Krishna D. Sharma, Bijay P. Chhetri, Charlette M. Parnell, Jennifer Yanhua Xie, Malathi Srivatsan, and Anindya Ghosh. 2021. "Functionalized Nanocellulose Drives Neural Stem Cells toward Neuronal Differentiation" Journal of Functional Biomaterials 12, no. 4: 64. https://doi.org/10.3390/jfb12040064