Hematological- and Immunological-Related Biomarkers to Characterize Patients with COVID-19 from Other Viral Respiratory Diseases
Abstract
:1. Introduction
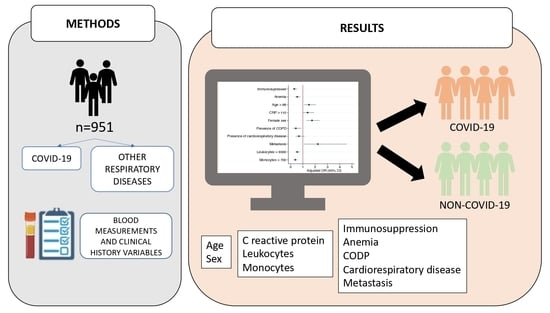
2. Materials and Methods
2.1. Study Design and Settings
2.2. Data Collection
2.2.1. Participants
2.2.2. Variables
2.3. Statistical Analyses
3. Results
| Variables | Total of Patients n = 951 | Univariate Analysis OR (CI) | p Value | Multivariate Analysis OR (CI) | p Value |
|---|---|---|---|---|---|
| Age > 65, % | 577 (60.7) | 1.17 (0.89–1.55) | 0.253 | 1.61 (1.16–2.23) | 0.004 |
| Sex male, % | 532 (55.9) | 1.56 (1.11–2.06) | 0.002 | 1.73 (1.27–2.36) | <0.001 |
| Oxygen saturation > 90%, % | 574 (60.4) | 1.41 (1.06–1.87) | 0.016 | 1.50 (1.09–2.07) | 0.012 |
| Monocytes < 500 cells/µL, % | 427 (44.9) | 2.95 (2.23–3.90) | <0.001 | 2.21 (1.59–3.08) | <0.001 |
| Neutrophils < 5500 cells/µL, % | 445 (46.8) | 1.72 (1.31–2.27) | <0.001 | 1.53 (1.09–2.14) | 0.013 |
| Lymphocytes < 1500 cells/µL, % | 723 (76) | 2.31 (1.62–3.31) | <0.001 | 1.69 (1.13–2.52) | 0.01 |
| C-reactive protein > 80 mg/dL, % | 450 (47.3) | 1.46 (1.11–1.91) | 0.006 | 1.55 (1.12–2.16) | 0.008 |
| Onset of symptoms > 4 días, % | 473 (49.7) | 2.05 (1.56–2.71) | <0.001 | 1.46 (1.07–1.99) | 0.015 |
| Anemia, % | 315 (33.1) | 0.47 (0.34–0.64) | <0.001 | 0.56 (0.38–0.38) | 0.004 |
| Cardiorespiratory disease, % | 216 (22.7) | 0.44 (0.30–0.63) | <0.001 | 0.47 (0.31–0.71) | <0.001 |
| Immunosuppression, % | 167 (17.6) | 0.33 (0.21–0.52) | <0.001 | 0.37 (0.22–0.61) | <0.001 |

4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tay, M.Z.; Poh, C.M.; Rénia, L.; MacAry, P.A.; Ng, L.F.P. The trinity of COVID-19: Immunity, inflammation and intervention. Nat. Rev. Immunol. 2020, 20, 363–374. [Google Scholar] [CrossRef]
- Waghmode, R.; Jadhav, S.; Nema, V. The Burden of Respiratory Viruses and Their Prevalence in Different Geographical Regions of India: 1970–2020. Front. Microbiol. 2021, 12, 2432. [Google Scholar] [CrossRef]
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2020, 19, 141–154. [Google Scholar] [CrossRef]
- Ticona, J.H.; Zaccone, V.M.; McFarlane, I.M. Community-Acquired Pneumonia: A Focused Review. Am. J. Med. Case Rep. 2021, 9, 45. [Google Scholar] [CrossRef]
- Pagliano, P.; Scarpati, G.; Sellitto, C.; Conti, V.; Spera, A.M.; Ascione, T.; Piazza, O.; Filippelli, A. Experimental Pharmacotherapy for COVID-19: The Latest Advances. J. Exp. Pharmacol. 2021, 13, 1–13. [Google Scholar] [CrossRef]
- De Almeida-Pititto, B.; Dualib, P.M.; Zajdenverg, L.; Dantas, J.R.; De Souza, F.D.; Rodacki, M.; Bertoluci, M.C. Severity and mortality of COVID 19 in patients with diabetes, hypertension and cardiovascular disease: A meta-analysis. Diabetol. Metab. Syndr. 2020, 12, 75. [Google Scholar] [CrossRef]
- Zabetakis, I.; Lordan, R.; Norton, C.; Tsoupras, A. COVID-19: The Inflammation Link and the Role of Nutrition in Potential Mitigation. Nutrients 2020, 12, 1466. [Google Scholar] [CrossRef]
- Anderson, R.M.; Vegvari, C.; Hollingsworth, T.D.; Pi, L.; Maddren, R.; Ng, C.W.; Baggaley, R.F. The SARS-CoV-2 pandemic: Remaining uncertainties in our understanding of the epidemiology and transmission dynamics of the virus, and challenges to be overcome. Interface Focus 2021, 11, 20210008. [Google Scholar] [CrossRef]
- Zheng, J. SARS-CoV-2: An Emerging Coronavirus that Causes a Global Threat. Int. J. Biol. Sci. 2020, 16, 1678. [Google Scholar] [CrossRef] [Green Version]
- Song, L.; Liang, E.Y.; Wang, H.M.; Shen, Y.; Kang, C.M.; Xiong, Y.J.; He, M.; Fu, W.J.; Ke, P.F.; Huang, X.Z. Differential diagnosis and prospective grading of COVID-19 at the early stage with simple hematological and biochemical variables. Diagn. Microbiol. Infect. Dis. 2021, 99, 115169. [Google Scholar] [CrossRef]
- Roberts, M.; Driggs, D.; Thorpe, M.; Gilbey, J.; Yeung, M.; Ursprung, S.; Aviles-Rivero, A.I.; Etmann, C.; McCague, C.; Beer, L.; et al. Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nat. Mach. Intell. 2021, 3, 199–217. [Google Scholar] [CrossRef]
- Lun, H.; Yang, W.; Zhao, S.; Jiang, M.; Xu, M.; Liu, F.; Wang, Y. Altered gut microbiota and microbial biomarkers associated with chronic kidney disease. Microbiologyopen 2019, 8, e00678. [Google Scholar] [CrossRef]
- Quiroz-Juárez, M.A.; Torres-Gómez, A.; Hoyo-Ulloa, I.; de León-Montiel, R.D.J.; U’Ren, A.B. Identification of high-risk COVID-19 patients using machine learning. PLoS ONE 2021, 16, e0257234. [Google Scholar] [CrossRef]
- Niemi, M.E.K.; Daly, M.J.; Ganna, A. The human genetic epidemiology of COVID-19. Nat. Rev. Genet. 2022, 1–14. [Google Scholar] [CrossRef]
- Gupta, A.; Madhavan, M.V.; Sehgal, K.; Nair, N.; Mahajan, S.; Sehrawat, T.S.; Bikdeli, B.; Ahluwalia, N.; Ausiello, J.C.; Wan, E.Y.; et al. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020, 26, 1017–1032. [Google Scholar] [CrossRef]
- Vargas-Mendoza, N.; García-Machorro, J.; Angeles-Valencia, M.; Martínez-Archundia, M.; Madrigal-Santillán, E.O.; Morales-González, Á.; Anguiano-Robledo, L.; Morales-González, J.A. Liver disorders in COVID-19, nutritional approaches and the use of phytochemicals. World J. Gastroenterol. 2021, 27, 5630. [Google Scholar] [CrossRef]
- Rendeiro, A.F.; Vorkas, C.K.; Krumsiek, J.; Singh, H.K.; Kapadia, S.N.; Cappelli, L.V.; Cacciapuoti, M.T.; Inghirami, G.; Elemento, O.; Salvatore, M. Metabolic and Immune Markers for Precise Monitoring of COVID-19 Severity and Treatment. Front. Immunol. 2022, 12, 809937. [Google Scholar] [CrossRef]
- Bowring, M.G.; Wang, Z.; Xu, Y.; Betz, J.; Muschelli, J.; Garibaldi, B.T.; Zeger, S.L. Outcome-Stratified Analysis of Biomarker Trajectories for Patients Infected with Severe Acute Respiratory Syndrome Coronavirus 2. Am. J. Epidemiol. 2021, 190, 2094–2106. [Google Scholar] [CrossRef]
- Cook, R.J.; Dickens, B.M.; Fathalla, M.F. World Medical Association Declaration of Helsinki: Ethical principles for medical research involving human subjects. J. Am. Coll. Dent. 2014, 81, 14–18. [Google Scholar] [CrossRef] [Green Version]
- Anemia. Available online: https://www.who.int/es/health-topics/anaemia#tab=tab_1 (accessed on 10 June 2022).
- Charlson, M.; Szatrowski, T.P.; Peterson, J.; Gold, J. Validation of a combined comorbidity index. J. Clin. Epidemiol. 1994, 47, 1245–1251. [Google Scholar] [CrossRef]
- Sundararajan, V.; Henderson, T.; Perry, C.; Muggivan, A.; Quan, H.; Ghali, W.A. New ICD-10 version of the Charlson comorbidity index predicted in-hospital mortality. J. Clin. Epidemiol. 2004, 57, 1288–1294. [Google Scholar] [CrossRef]
- Lindsey, C. VSELECT: Stata module to perform linear regression variable selection. In Statistical Software Components; Boston College: Chestnut Hill, MA, USA, 2014. [Google Scholar]
- Xiang, Q.; Feng, Z.; Diao, B.; Qinghua, Q.; Yang, H.; Zhang, Y.; Wang, G.; Wang, H.; Wang, C.; Liu, L.; et al. SARS-CoV-2 Induces Lympohcytopenia by Promoting Immaflamation and Decimates Secondary Lymphoid Organs. Front Immunol. 2021, 12, 661052. [Google Scholar] [CrossRef]
- Wilder-Smith, A. COVID-19 in comparison with other emerging viral diseases: Risk of geographic spread via travel. Trop. Dis. Travel Med. Vaccines 2021, 7, 3. [Google Scholar] [CrossRef]
- Richardson, S.; Hirsch, J.S.; Narasimhan, M.; Crawford, J.M.; McGinn, T.; Davidson, K.W.; Barnaby, D.P.; Becker, L.B.; Chelico, J.D.; Cohen, S.L.; et al. Presenting Characteristics, Comorbidities, and Outcomes Among 5700 Patients Hospitalized With COVID-19 in the New York City Area. JAMA 2020, 323, 2052–2059. [Google Scholar] [CrossRef]
- Knight, S.R.; Ho, A.; Pius, R.; Buchan, I.; Carson, G.; Drake, T.M.; Dunning, J.; Fairfield, C.J.; Gamble, C.; Green, C.A.; et al. Risk stratification of patients admitted to hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: Development and validation of the 4C Mortality Score. BMJ 2020, 370, 22. [Google Scholar] [CrossRef]
- Yavuz, B.G.; Colak, S.; Guven, R.; Altundag, İ.; Seyhan, A.U.; Inanc, R.G. Clinical Features of the 60 Years and Older Patients Infected with 2019 Novel Coronavirus: Can We Predict Mortality Earlier? Gerontology 2021, 67, 433–440. [Google Scholar] [CrossRef]
- Flaherty, G.T.; Hession, P.; Liew, C.H.; Lim, B.C.W.; Leong, T.K.; Lim, V.; Sulaiman, L.H. COVID-19 in adult patients with pre-existing chronic cardiac, respiratory and metabolic disease: A critical literature review with clinical recommendations. Trop. Dis. Travel Med. Vaccines 2020, 6, 16. [Google Scholar] [CrossRef]
- KimID, Y.; Zhu, L.; Zhu, H.; Li, X.; Huang, Y.; Gu, C.; Bush, H.; Chung, C.; Zhang, G.-Q. Characterizing cancer and COVID-19 outcomes using electronic health records. PLoS ONE 2022, 17, e0267584. [Google Scholar] [CrossRef]
- Torres Acosta, M.A.; Singer, B.D. Pathogenesis of COVID-19-induced ARDS: Implications for an ageing population. Eur. Respir. J. 2020, 56, 2002049. [Google Scholar] [CrossRef]
- Tavakolpour, S.; Rakhshandehroo, T.; Wei, E.X.; Rashidian, M. Lymphopenia during the COVID-19 infection: What it shows and what can be learned. Immunol. Lett. 2020, 225, 31–32. [Google Scholar] [CrossRef]
- Ramos-Lopez, O.; San-Cristobal, R.; Martinez-Urbistondo, D.; Micó, V.; Colmenarejo, G.; Villares-Fernandez, P.; Daimiel, L.; Alfredo Martinez, J. Proinflammatory and hepatic features related to morbidity and fatal outcomes in covid-19 patients. J. Clin. Med. 2021, 10, 3112. [Google Scholar] [CrossRef]
- Prozan, L.; Shusterman, E.; Ablin, J.; Mitelpunkt, A.; Weiss-Meilik, A.; Adler, A.; Choshen, G.; Kehat, O. Prognostic value of neutrophil-to-lymphocyte ratio in COVID-19 compared with Influenza and respiratory syncytial virus infection. Sci. Rep. 2021, 11, 21519. [Google Scholar] [CrossRef]
- King, A.H.; Mehkri, O.; Rajendram, P.; Wang, X.; Vachharajani, V.; Duggal, A. A High Neutrophil-Lymphocyte Ratio Is Associated With Increased Morbidity and Mortality in Patients With Coronavirus Disease 2019. Crit. Care Explor. 2021, 3, e0444. [Google Scholar] [CrossRef]
- Yang, A.-P.; Liu, J.-P.; Tao, W.-Q.; Li, H.-M. The diagnostic and predictive role of NLR, d-NLR and PLR in COVID-19 patients. Int. Immunopharmacol. 2020, 84, 106504. [Google Scholar] [CrossRef]
- Citu, C.; Gorun, F.; Motoc, A.; Sas, I.; Gorun, O.M.; Burlea, B.; Tuta-Sas, I.; Tomescu, L.; Neamtu, R.; Malita, D.; et al. The Predictive Role of NLR, d-NLR, MLR, and SIRI in COVID-19 Mortality. Diagnostics 2022, 12, 122. [Google Scholar] [CrossRef]
- Sacco, K.; Castagnoli, R.; Vakkilainen, S.; Liu, C.; Delmonte, O.M.; Oguz, C.; Kaplan, I.M.; Alehashemi, S.; Burbelo, P.D.; Bhuyan, F.; et al. Immunopathological signatures in multisystem inflammatory syndrome in children and pediatric COVID-19. Nat. Med. 2022, 28, 1050–1062. [Google Scholar] [CrossRef]
- Mohiuddin, M.; Kasahara, K. Cancer metastasis may increase COVID-19 mortality: Suitable targets required to impede cancer metastasis. J. Infect. Public Health 2022, 15, 153. [Google Scholar] [CrossRef]
- Tahery, N.; Khodadost, M.; Sherafat, S.J.; Tavirani, M.R.; Ahmadi, N.; Montazer, F.; Tavirani, M.R.; Naderi, N. C-reactive protein as a possible marker for severity and mortality of COVID-19 infection. Gastroenterol. Hepatol. Bed Bench 2021, 14, S118. [Google Scholar] [CrossRef]
- Liu, Y.; Mao, B.; Liang, S.; Yang, J.W.; Lu, H.W.; Chai, Y.H.; Wang, L.; Zhang, L.; Li, Q.H.; Zhao, L.; et al. Association between age and clinical characteristics and outcomes of COVID-19. Eur. Respir. J. 2020, 55, 2001112. [Google Scholar] [CrossRef] [Green Version]
- Jacobsen, H.; Klein, S.L. Sex Differences in Immunity to Viral Infections. Front. Immunol. 2021, 12, 3483. [Google Scholar] [CrossRef]
- Parimoo, A.; Biswas, A.; Baitha, U.; Gupta, G.; Pandey, S.; Ranjan, P.; Gupta, V.; Roy, D.B.; Prakash, B.; Wig, N. Dynamics of Inflammatory Markers in Predicting Mortality in COVID-19. Cureus 2021, 13, e19080. [Google Scholar] [CrossRef]
- Citu, C.; Burlea, B.; Gorun, F.; Motoc, A.; Gorun, O.M.; Malita, D.; Ratiu, A.; Margan, R.; Grigoras, M.L.; Bratosin, F.; et al. Predictive Value of Blood Coagulation Parameters in Poor Outcomes in COVID-19 Patients: A Retrospective Observational Study in Romania. J. Clin. Med. 2022, 11, 2831. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Huang, J.; Li, C.; Zhao, Y.; Wang, D.; Huang, Z.; Yang, K. The role of seasonality in the spread of COVID-19 pandemic. Environ. Res. 2021, 195, 110874. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.H. Sample sizes in COVID-19–related research. Can. Med. Assoc. J. 2020, 192, E461. [Google Scholar] [CrossRef]


| Variable | COVID-19 Positive n = 313 | COVID-19 Negative n = 237 | Flu Positive n = 119 | Pan-Viral Positive n = 116 | Pan-Viral Negative n = 166 | Global n = 951 | p Value |
|---|---|---|---|---|---|---|---|
| Age (y) | 69.5 (14.7) | 69.5 (16.3) | 66.5 (17.1) | 66.9 (17.1) | 66.3 (17.1) | 68.2 (16.2) | 0.256 |
| Male sex (%) | 198 (63.3) | 139 (58.6) | 58 (48.7) | 51 (44.0) | 86 (51.8) | 532 (55.9) | 0.001 |
| Cardiovascular disease (%) | 33 (10.5) | 31 (13.1) | 22 (18.5) | 26 (22.4) | 33 (11.4) | (8.0) | 0.593 |
| Cerebrovascular disease (%) | 18 (5.7) | 14 (5.9) | 2 (2.5) | 3 (2.9) | 11 (6.6) | 48 (5.1) | 0.823 |
| Cardiorespiratory disease (%) | 44 (14.0) | 49 (20.6) | 32 (26.8) | 73 (62.9) | 50 (30.1) | 248 (26.1) | 0.027 |
| Chronic obstructive pulmonary disease (%) | 18 (5.8) | 29 (12.2) | 22 (18.5) | 26 (22.4) | 30 (18.1) | 125 (13.1) | <0.001 |
| Diabetes Mellitus (%) | 55 (17.6) | 40 (16.9) | 13 (10.9) | 16 (13.8) | 15 (9) | 139 (14.6) | 0.277 |
| Chronic hepatic disease (%) | 2 (0.6) | 2 (0.8) | 0 (0) | 1 (0.8) | 5 (3.0) | 10 (1.1) | 0.866 |
| Dementia (%) | 21 (6.7) | 23 (9.7) | 1 (0.8) | 3 (2.6) | 4 (2.4) | 52 (5.5) | 0.031 |
| Connective tissue disease (%) | 6 (1.9) | 5 (2.1) | 5 (4.2) | 5 (4.3) | 15 (9) | 36 (3.8) | 0.197 |
| Chronic kidney disease (%) | 10 (3.2) | 9 (3.8) | 5 (4.2) | 4 (3.4) | 5 (3) | 33 (3.5) | 0.967 |
| Acquired Immuno Deficiency Syndrom (AIDS) (%) | 0 (0) | 1 (0.4) | 0 (0) | 1 (0.8) | 0 (0) | 2 (0.2) | 0.624 |
| Leukemia (%) | 5 (1.6) | 5 (2.1) | 2 (1.7) | 7 (6.0) | 25 (15.1) | 44 (4.6) | <0.001 |
| Lymphoma (%) | 11 (3.5) | 8 (3.4) | 7 (5.9) | 4 (3.4) | 16 (9.6) | 46 (4.8) | 0.384 |
| Solid tumor (%) | 41 (13.1) | 19 (8) | 13 (10.9) | 13 (11.2) | 23 (13.9) | 109 (11.5) | 0.73 |
| Metastatic tumor (%) | 17 (5.4) | 13 (5.5) | 11 (9.2) | 10 (8.6) | 11 (6.6) | 62 (6.5) | 0.375 |
| Anemia (%) | 71 (22.7) | 54 (23.1) | 52 (43.3) | 33 (28.4) | 16 (9.6) | 226 (23.7) | 0.001 |
| Hemiplegia (%) | 0 (0) | 3 (1.2) | 0 (0) | 0 (0) | 0 (0) | 3 (0.3) | 0.322 |
| Onset symptons (days) | 6.4 (6.23) | 6.8 (5.4) | 4.8 (5.52) | 5.8 (6.22) | 5.4 (5.25) | 6.1 (5.81) | 0.095 |
| Hospitalization (days) | 12.2 (11.9) | 9.4 (9.7) | 11.3 (9.8) | 11.2 (11.9) | 14.8 (19.9) | 11.7 (13.1) | 0.850 |
| Inmunosupression (%) | 27 (8.6) | 19 (8.0) | 24 (20.2) | 28 (24.1) | 69 (41.6) | 167 (17.6) | <0.001 |
| Exitus (%) | 71 (22.7) | 24 (10.1) | 7 (5.9) | 4 (3.4) | 15 (9) | 121 (12.7) | <0.001 |
| Variables (at the Moment of Hospitalization) | COVID-19 Positive n = 313 | COVID-19 Negative n = 237 | Flu Positive n = 119 | Pan-Viral Positive n = 116 | Pan-Viral Negative n = 166 | Global n = 951 | p Value * |
|---|---|---|---|---|---|---|---|
| Hemoglobin (g/dL) | 13.56 (1.99) | 13.61 (2.01) | 12.55 (2.44) | 12.42 (2.33) | 11.96 (2.48) | 13.03 (2.29) | <0.001 |
| Leukocytes (cells/µL) | 7.703 (4.261) | 9.010 (4.847) | 10.287 (8.986) | 9.873 (6.937) | 10.806 (9.260) | 9.162 (6.635) | <0.001 |
| Neutrophils, (cells/µL) | 6.199 (4.089) | 6.941 (4.318) | 7.630 (5.692) | 8.046 (6.517) | 7.734 (5.640) | 7.055 (5.025) | 0.001 |
| Lymphocytes (cells/µL) | 961 (497) | 1.295 (997) | 1.420 (3.103) | 1.106 (976) | 1.695 (4.333) | 1.249 (2.237) | 0.020 |
| Monocytes (cells/µL) | 491 (434) | 654 (415) | 979 (2.845) | 696 (696) | 1.244 (4.846) | 750 (2.312) | 0.084 |
| C-reactive protein (mg/dL) | 120.52 (105.61) | 95.38 (99.86) | 125.67 (130.91) | 110.5 (127.10) | 115.22 (120.24) | 112.73 (111.41) | 0.116 |
| Neutrophils/Lymphocytes ratio | 8.75 (1.3) | 7.88 (0.7) | 9.32 (1.6) | 9.86 (2.1) | 9.11 (2.3) | 9.49 (2.6) | <0.001 |
| OR | 95% CI | p Value | AUC | |
|---|---|---|---|---|
| MODEL (R2 = 0.13) | 0.75 | |||
| Age > 68 (y) | 1.49 | (1.08–2.06) | <0.001 | |
| Sex female | 1.74 | (1.28–2.37) | <0.001 | |
| Anemia | 0.49 | (0.37–0.79) | 0.002 | |
| Immunosuppression | 0.31 | (0.17–0.54) | <0.001 | |
| C-reactive protein > 110 (mg/L) | 1.39 | (1.00–1.92) | 0.047 | |
| CODP | 0.44 | (0.21–0.87) | 0.020 | |
| Cardiorespiratory disease | 0.68 | (0.40–1.15) | 0.156 | |
| Metastasis | 2.22 | (1.07–4.57) | 0.031 | |
| Leukocytes > 9000 (cells/µL) | 0.54 | (0.37–0.76) | 0.001 | |
| Monocytes > 700 (cells/µL) | 0.38 | (0.26–0.55) | <0.001 |
| Variable (Cells/µL) | COVID-19 Negative (n = 472) | COVID-19 Positive (n = 313) | p Value |
|---|---|---|---|
| Leukocytes | 9811 ± 7443 | 7703 ± 4261 | <0.001 |
| Neutrophils | 7379 ± 5408 | 6199 ± 4089 | 0.002 |
| Monocytes | 855 ± 2766 | 491 ± 434 | 0.016 |
| Lymphocytes | 1370 ± 2684 | 961 ± 497 | 0.005 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suárez-Del-Villar-Carrero, R.; Martinez-Urbistondo, D.; Cuevas-Sierra, A.; Ibañez-Sustacha, I.; Candela-Fernandez, A.; Dominguez-Calvo, A.; Ramos-Lopez, O.; Vargas, J.A.; Reglero, G.; Villares-Fernandez, P.; et al. Hematological- and Immunological-Related Biomarkers to Characterize Patients with COVID-19 from Other Viral Respiratory Diseases. J. Clin. Med. 2022, 11, 3578. https://doi.org/10.3390/jcm11133578
Suárez-Del-Villar-Carrero R, Martinez-Urbistondo D, Cuevas-Sierra A, Ibañez-Sustacha I, Candela-Fernandez A, Dominguez-Calvo A, Ramos-Lopez O, Vargas JA, Reglero G, Villares-Fernandez P, et al. Hematological- and Immunological-Related Biomarkers to Characterize Patients with COVID-19 from Other Viral Respiratory Diseases. Journal of Clinical Medicine. 2022; 11(13):3578. https://doi.org/10.3390/jcm11133578
Chicago/Turabian StyleSuárez-Del-Villar-Carrero, Rafael, Diego Martinez-Urbistondo, Amanda Cuevas-Sierra, Iciar Ibañez-Sustacha, Alberto Candela-Fernandez, Andrea Dominguez-Calvo, Omar Ramos-Lopez, Juan Antonio Vargas, Guillermo Reglero, Paula Villares-Fernandez, and et al. 2022. "Hematological- and Immunological-Related Biomarkers to Characterize Patients with COVID-19 from Other Viral Respiratory Diseases" Journal of Clinical Medicine 11, no. 13: 3578. https://doi.org/10.3390/jcm11133578