Identification and Expression of Inward-Rectifying Potassium Channel Subunits in Plutella xylostella
Abstract
:1. Introduction
2. Materials and Methods
2.1. Insects and Tissue Isolation
2.2. RNA Isolation, cDNA Preparation, and TA Cloning
2.3. Sequences Alignment and Phylogenetic Analysis
2.4. Quantitative Real-Time PCR
2.5. Data Analysis and Statistics
3. Results
3.1. Kir Channel Subunits in Lepidoptera
3.2. The pxkirs Clone
3.3. Sequence Analysis
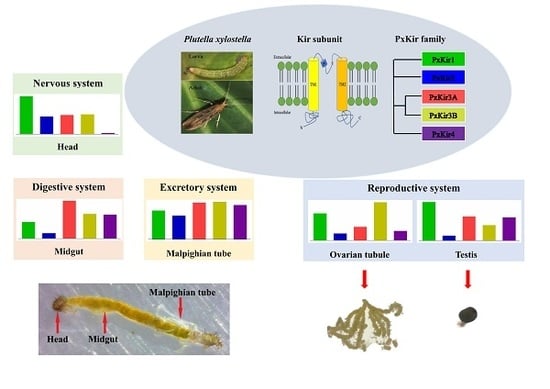
3.4. Developmental and Tissue Distribution of kirs in P. xylostella
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Cycle Steps | pxkir1 | pxkir2 | pxkir3a | pxkir3b | pxkir4 | Cycles |
|---|---|---|---|---|---|---|
| Initial denaturation | 95 °C 3 min | 95 °C 3 min | 95 °C 3 min | 95 °C 3 min | 95 °C 3 min | 1 |
| Denaturation | 95 °C 15 s | 95 °C 15 s | 95 °C 15 s | 95 °C 15 s | 95 °C 15 s | 32 |
| Annealing | 60 °C 15 s | 50 °C 15 s | 62 °C 15 s | 58 °C 15 s | 60 °C 15 s | |
| Extension | 72 °C 120 s | 72 °C 90 s | 72 °C 100 s | 72 °C 100 s | 72 °C 90 s | |
| Final elongation | 72 °C 5 min | 72 °C 5 min | 72 °C 5 min | 72 °C 5 min | 72 °C 5 min | 1 |
| Kir of P. xylostella | Kir of Other Insects | Similarity (%) |
|---|---|---|
| PxKir1 | AaKir1 | 52 |
| AgKir1 | 54 | |
| DmKir1 | 52 | |
| NlKir1 | 67 | |
| ApKir1 | 61 | |
| PxKir2 | AaKir2A | 77 |
| AaKir2B | 64 | |
| AaKir2B’ | 64 | |
| AgKir2A | 74 | |
| AgKir2A’ | 67 | |
| AgKir2B | 61 | |
| DmKir2 | 69 | |
| NlKir2 | 67 | |
| ApKir2 | 55 | |
| PxKir3A | AaKir3 | 33 |
| AgKir3A | 39 | |
| DmKir3 | 34 | |
| NlKir3 | 32 | |
| PxKir3B | AaKir3 | 39 |
| AgKir3A | 38 | |
| DmKir3 | 38 | |
| NlKir3 | 36 | |
| PxKir4 | BmKir4A | 61 |
| BmKir4B | 47 | |
| MsKir4A | 59 | |
| MsKir4B | 51 | |
| DpKir4 | 47 |
References
- Minor, D.L., Jr.; Masseling, S.J.; Jan, Y.N.; Jan, L.Y. Transmembrane structure of an inwardly rectifying potassium channel. Cell 1999, 96, 879–891. [Google Scholar] [CrossRef] [Green Version]
- Lu, Z.; Klem, A.M.; Ramu, Y. Ion conduction pore is conserved among potassium channels. Nature 2001, 413, 809–813. [Google Scholar] [CrossRef] [PubMed]
- Hibino, H.; Inanobe, A.; Furutani, K.; Murakami, S.; Findlay, I.; Kurachi, Y. Inwardly rectifying potassium channels: Their structure, function, and physiological roles. Physiol. Rev. 2010, 90, 291–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez, C.; Baez-Nieto, D.; Valencia, I.; Oyarzun, I.; Rojas, P.; Naranjo, D.; Latorre, R. K+ channels: Function-structural overview. Compr. Physiol. 2012, 2, 2087–2149. [Google Scholar] [CrossRef]
- Abraham, M.R.; Jahangir, A.; Alekseev, A.E.; Terzic, A. Channelopathies of inwardly rectifying potassium channels. FASEB J. 1999, 13, 1901–1910. [Google Scholar] [CrossRef] [PubMed]
- Pattnaik, B.R.; Asuma, M.P.; Spott, R.; Pillers, D.A. Genetic defects in the hotspot of inwardly rectifying K+ (Kir) channels and their metabolic consequences: A review. Mol. Genet. Metab. 2012, 105, 64–72. [Google Scholar] [CrossRef] [Green Version]
- Mayfield, J.; Blednov, Y.A.; Harris, R.A. Behavioral and Genetic Evidence for GIRK Channels in the CNS: Role in Physiology, Pathophysiology, and Drug Addiction. Int. Rev. Neurobiol. 2015, 123, 279–313. [Google Scholar] [CrossRef] [Green Version]
- Sacco, S.; Giuliano, S.; Sacconi, S.; Desnuelle, C.; Barhanin, J.; Amri, E.Z.; Bendahhou, S. The inward rectifier potassium channel Kir2.1 is required for osteoblastogenesis. Hum. Mol. Genet. 2015, 24, 471–479. [Google Scholar] [CrossRef] [Green Version]
- Tinker, A.; Aziz, Q.; Li, Y.W.; Specterman, M. ATP-Sensitive Potassium Channels and Their Physiological and Pathophysiological Roles. Compr. Physiol. 2018, 8, 1463–1511. [Google Scholar] [CrossRef]
- Clarke, O.B.; Caputo, A.T.; Hill, A.P.; Vandenberg, J.I.; Smith, B.J.; Gulbis, J.M. Domain reorientation and rotation of an intracellular assembly regulate conduction in Kir potassium channels. Cell 2010, 141, 1018–1029. [Google Scholar] [CrossRef] [Green Version]
- Li, N.N.; Wu, J.X.; Ding, D.; Cheng, J.X.; Gao, N.; Chen, L. Structure of a Pancreatic ATP-Sensitive Potassium Channel. Cell 2017, 168, 101–110.e110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, X.; Avalos, J.L.; Chen, J.Y.; MacKinnon, R. Crystal structure of the eukaryotic strong inward-rectifier K+ channel Kir2.2 at 3.1 A resolution. Science 2009, 326, 1668–1674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, S.B.; Tao, X.; MacKinnon, R. Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2. Nature 2011, 477, 495–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishida, M.; MacKinnon, R. Structural basis of inward rectification: Cytoplasmic pore of the G protein-gated inward rectifier GIRK1 at 1.8 A resolution. Cell 2002, 111, 957–965. [Google Scholar] [CrossRef] [Green Version]
- Rouhier, M.F.; Raphemot, R.; Denton, J.S.; Piermarini, P.M. Pharmacological validation of an inward-rectifier potassium (Kir) channel as an insecticide target in the yellow fever mosquito Aedes aegypti. PLoS ONE 2014, 9, e100700. [Google Scholar] [CrossRef]
- Reale, V.; Chatwin, H.M.; Evans, P.D. The activation of G-protein gated inwardly rectifying K+ channels by a cloned Drosophila melanogaster neuropeptide F-like receptor. Eur. J. Neurosci. 2004, 19, 570–576. [Google Scholar] [CrossRef]
- Piermarini, P.M.; Dunemann, S.M.; Rouhier, M.F.; Calkins, T.L.; Raphemot, R.; Denton, J.S.; Hine, R.M.; Beyenbach, K.W. Localization and role of inward rectifier K+ channels in Malpighian tubules of the yellow fever mosquito Aedes aegypti. Insect Biochem. Mol. Biol. 2015, 67, 59–73. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.X.; Statler, B.M.; Calkins, T.L.; Alfaro, E.; Esquivel, C.J.; Rouhier, M.F.; Denton, J.S.; Piermarini, P.M. Dynamic expression of genes encoding subunits of inward rectifier potassium (Kir) channels in the yellow fever mosquito Aedes aegypti. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2017, 204, 35–44. [Google Scholar] [CrossRef]
- Ren, M.M.; Niu, J.G.; Hu, B.; Wei, Q.; Zheng, C.; Tian, X.R.; Gao, C.F.; He, B.J.; Dong, K.; Su, J.Y. Block of Kir channels by flonicamid disrupts salivary and renal excretion of insect pests. Insect Biochem. Mol. Biol. 2018, 99, 17–26. [Google Scholar] [CrossRef]
- Doring, F.; Wischmeyer, E.; Kuhnlein, R.P.; Jackle, H.; Karschin, A. Inwardly rectifying K+ (Kir) channels in Drosophila. A crucial role of cellular milieu factors Kir channel function. J. Biol. Chem. 2002, 277, 25554–25561. [Google Scholar] [CrossRef] [Green Version]
- MacLean, S.J.; Andrews, B.C.; Verheyen, E.M. Characterization of Dir: A putative potassium inward rectifying channel in Drosophila. Mech. Dev. 2002, 116, 193–197. [Google Scholar] [CrossRef]
- Piermarini, P.M.; Rouhier, M.F.; Schepel, M.; Kosse, C.; Beyenbach, K.W. Cloning and functional characterization of inward-rectifying potassium (Kir) channels from Malpighian tubules of the mosquito Aedes aegypti. Insect Biochem. Mol. Biol. 2013, 43, 75–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piermarini, P.M.; Inocente, E.A.; Acosta, N.; Hopkins, C.R.; Denton, J.S.; Michel, A.P. Inward rectifier potassium (Kir) channels in the soybean aphid Aphis glycines: Functional characterization, pharmacology, and toxicology. J. Insect Physiol. 2018, 110, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Raphemot, R.; Estevez-Lao, T.Y.; Rouhier, M.F.; Piermarini, P.M.; Denton, J.S.; Hillyer, J.F. Molecular and functional characterization of Anopheles gambiae inward rectifier potassium (Kir1) channels: A novel role in egg production. Insect Biochem. Mol. Biol. 2014, 51, 10–19. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.P.; Baum, M.; Huang, C.L.; Rodan, A.R. Two inwardly rectifying potassium channels, Irk1 and Irk2, play redundant roles in Drosophila renal tubule function. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2015, 309, R747–R756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piermarini, P.M.; Esquivel, C.J.; Denton, J.S. Malpighian Tubules as Novel Targets for Mosquito Control. Int. J. Environ. Res. Public Health 2017, 14, 111. [Google Scholar] [CrossRef] [Green Version]
- Lewis, L.M.; Bhave, G.; Chauder, B.A.; Banerjee, S.; Lornsen, K.A.; Redha, R.; Fallen, K.; Lindsley, C.W.; Weaver, C.D.; Denton, J.S. High-throughput screening reveals a small-molecule inhibitor of the renal outer medullary potassium channel and Kir7.1. Mol. Pharmacol. 2009, 76, 1094–1103. [Google Scholar] [CrossRef] [Green Version]
- Kharade, S.V.; Sheehan, J.H.; Figueroa, E.E.; Meiler, J.; Denton, J.S. Pore Polarity and Charge Determine Differential Block of Kir1.1 and Kir7.1 Potassium Channels by Small-Molecule Inhibitor VU590. Mol. Pharmacol. 2017, 92, 338–346. [Google Scholar] [CrossRef]
- Swale, D.R.; Li, Z.L.; Guerrero, F.; Perez De Leon, A.A.; Foil, L.D. Role of inward rectifier potassium channels in salivary gland function and sugar feeding of the fruit fly, Drosophila melanogaster. Pestic. Biochem. Physiol. 2017, 141, 41–49. [Google Scholar] [CrossRef] [Green Version]
- Dahal, G.R.; Pradhan, S.J.; Bates, E.A. Inwardly rectifying potassium channels influence Drosophila wing morphogenesis by regulating Dpp release. Development 2017, 144, 2771–2783. [Google Scholar] [CrossRef] [Green Version]
- Fenner, K.; Canonica, S.; Wackett, L.P.; Elsner, M. Evaluating pesticide degradation in the environment: Blind spots and emerging opportunities. Science 2013, 341, 752–758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.Z.; Zhang, X.L.; Shen, J.; Li, D.Y.; Wan, H.; You, H.; Li, J.H. Cross-resistance and biochemical mechanisms of resistance to indoxacarb in the diamondback moth, Plutella xylostella. Pestic. Biochem. Physiol. 2017, 140, 85–89. [Google Scholar] [CrossRef]
- Yin, C.Y.; Wang, R.; Luo, C.; Zhao, K.; Wu, Q.Y.; Wang, Z.Y.; Yang, G.F. Monitoring, Cross-Resistance, Inheritance, and Synergism of Plutella xylostella (Lepidoptera: Plutellidae) Resistance to Pyridalyl in China. J. Econ. Entomol. 2019, 112, 329–334. [Google Scholar] [CrossRef]
- Xu, J.; Wang, Z.Y.; Wang, Y.F.; Ma, H.H.; Zhu, H.; Liu, J.; Zhou, Y.; Deng, X.L.; Zhou, X.M. ABCC2 participates in the resistance of Plutella xylostella to chemical insecticides. Pestic. Biochem. Physiol. 2020, 162, 52–59. [Google Scholar] [CrossRef]
- Ma, H.H.; Huang, Q.T.; Lai, X.Y.; Liu, J.; Zhu, H.; Zhou, Y.; Deng, X.L.; Zhou, X.M. Pharmacological Properties of the Type 1 Tyramine Receptor in the Diamondback Moth, Plutella xylostella. Int. J. Mol. Sci. 2019, 20, 2953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.L.; Guo, Z.J.; Kang, S.; Qin, J.Y.; Gong, L.J.; Sun, D.; Guo, L.; Zhu, L.H.; Bai, Y.; Zhang, Z.Z.; et al. Reduced expression of the P-glycoprotein gene PxABCB1 is linked to resistance to Bacillus thuringiensis Cry1Ac toxin in Plutella xylostella (L.). Pest Manag. Sci. 2020, 76, 712–720. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.T.; Ma, H.H.; Deng, X.L.; Zhu, H.; Liu, J.; Zhou, Y.; Zhou, X.M. Pharmacological characterization of a beta-adrenergic-like octopamine receptor in Plutella xylostella. Arch. Insect Biochem. Physiol. 2018, 98, e21466. [Google Scholar] [CrossRef]
- Fu, W.; Xie, W.; Zhang, Z.; Wang, S.L.; Wu, Q.J.; Liu, Y.; Zhou, X.; Zhou, X.M.; Zhang, Y.J. Exploring valid reference genes for quantitative real-time PCR analysis in Plutella xylostella (Lepidoptera: Plutellidae). Int. J. Biol. Sci. 2013, 9, 792–802. [Google Scholar] [CrossRef] [Green Version]
- You, Y.C.; Xie, M.; Vasseur, L.; You, M.S. Selecting and validating reference genes for quantitative real-time PCR in Plutella xylostella (L.). Genome 2018, 61, 349–358. [Google Scholar] [CrossRef] [Green Version]
- Hua, J.F.; Zhang, S.; Cui, J.J.; Wang, D.J.; Wang, C.Y.; Luo, J.Y.; Lv, L.M.; Ma, Y. Functional characterizations of one odorant binding protein and three chemosensory proteins from Apolygus lucorum (Meyer-Dur) (Hemiptera: Miridae) legs. J. Insect Physiol. 2013, 59, 690–696. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. J. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, R.; Wang, X.J.; Kassianos, A.J.; Zuryn, S.; Roper, K.E.; Osborne, A.; Sampangi, S.; Francis, L.; Raghunath, V.; Healy, H. Laser capture microdissection and multiplex-tandem PCR analysis of proximal tubular epithelial cell signaling in human kidney disease. PLoS ONE 2014, 9, e87345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raphemot, R.; Rouhier, M.F.; Hopkins, C.R.; Gogliotti, R.D.; Lovell, K.M.; Hine, R.M.; Ghosalkar, D.; Longo, A.; Beyenbach, K.W.; Denton, J.S.; et al. Eliciting renal failure in mosquitoes with a small-molecule inhibitor of inward-rectifying potassium channels. PLoS ONE 2013, 8, e64905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Neal, S.T.; Swale, D.R.; Anderson, T.D. ATP-sensitive inwardly rectifying potassium channel regulation of viral infections in honeybees. Sci. Rep. 2017, 7, 8668. [Google Scholar] [CrossRef] [Green Version]
- Raphemot, R.; Rouhier, M.F.; Swale, D.R.; Days, E.; Weaver, C.D.; Lovell, K.M.; Konkel, L.C.; Engers, D.W.; Bollinger, S.R.; Hopkins, C.; et al. Discovery and characterization of a potent and selective inhibitor of Aedes aegypti inward rectifier potassium channels. PLoS ONE 2014, 9, e110772. [Google Scholar] [CrossRef]
- Chen, R.; Swale, D.R. Inwardly rectifying potassium (Kir) channels represent acritical ion conductance pathway in the nervous systems of insects. Sci. Rep. 2018, 8, 1617. [Google Scholar] [CrossRef]
- Mamidala, P.; Mittapelly, P.; Jones, S.C.; Piermarini, P.M.; Mittapalli, O. Molecular characterization of genes encoding inward rectifier potassium (Kir) channels in the bed bug (Cimex lectularius). Comp. Biochem. Physiol. Part B 2013, 164, 275–279. [Google Scholar] [CrossRef]
- Rouhier, M.F.; Piermarini, P.M. Identification of life-stage and tissue-specific splice variants of an inward rectifying potassium (Kir) channel in the yellow fever mosquito Aedes aegypti. Insect Biochem. Mol. Biol. 2014, 48, 91–99. [Google Scholar] [CrossRef]
- Evans, J.M.; Allan, A.K.; Davies, S.A.; Dow, J.A. Sulphonylurea sensitivity and enriched expression implicate inward rectifier K+ channels in Drosophila melanogaster renal function. J. Exp. Biol. 2005, 208, 3771–3783. [Google Scholar] [CrossRef] [Green Version]




| Name | Primer Sequences (5′-3′) | Gene | Site | Purpose |
|---|---|---|---|---|
| PxKir1_F | ATGAAGAGATGTCGGCCATGAAGAGAT | pxkir1 | −8 | DNA cloning |
| PxKir1_R | TACACTTCTGCGTCTGGTGCTGACG | 1861 | ||
| PxKir2_F | CTTTCGTATTTAGTATCTATGGTT | pxkir2 | −92 | |
| PxKir2_R | AATGAGATAGTTCGTAAAGTTGCT | 1830 | ||
| PxKir3A_F | TAAGTTGGACACATCATTGAGCTT | pxkir3A | −135 | |
| PxKir3A_R | GAAAATCAGTTTCCTACGTTGGATAGTT | 1997 | ||
| PxKir3B_F | CTGCTGTAGGCTGATGATGGTGAT | pxkir3B | −123 | |
| PxKir3B_R | AAGTCAAGAATCGTTTTGTTATGTG | 1890 | ||
| PxKir4_F | TTTACCGGCATGGCATTCTATTACG | pxkir4 | −9 | |
| PxKir4_R | GAAAATGAAGGTAGCAACTTTACAAATACA | 1375 | ||
| PxKir1-qF | GAGGTGCTGCCGTTCTAC | pxkir1 | 703 | Real-time Quantitative PCR |
| PxKir1-qR | TCGACTCGATCACTCCCT | 895 | ||
| PxKir2-qF | ATTGGACAGTTCACGATACATAGGAA | pxkir2 | 336 | |
| PxKir2-qR | CCGAGCCAGGAGAGGATGAAG | 575 | ||
| PxKir3A-qF | GGGGGTACCGCTTCAAGAATGTTAT | pxkir3A | 1441 | |
| PxKir3A-qR | GGAGGAACCCTGTGTGACTGAG | 1626 | ||
| PxKir3B-qF | ACAAGGCTGGCTATGGCGACTA | pxkir3B | 610 | |
| PxKir3B-qR | GATGTACCAGAAACCAGCGAAC | 822 | ||
| PxKir4-qF | TATCATCCAGGAAGTGGCTAAGGCT | pxkir4 | 62 | |
| PxKir4-qR | GTCGGGAAACGCATGGTACTTGTGT | 234 | ||
| RPL8-qF | CGGTCGTGCCTACCACAAATACA | rpL8 | 559 | |
| RPL8-qR | CGTGAGGATGCTCCACAGGGT | 648 | ||
| RPS4-qF | ATGGATGTTGTGTCGATTGAAAAGA | rpS4 | 335 | |
| RPS4-qR | GAGTGATGCGGTGGATGGTGA | 423 | ||
| EF1-qF | GCCTCCCTACAGCGAATC | ef1 | 374 | |
| EF1-qR | CCTTGAACCAGGGCATCT | 535 |
| Gene | P. xylostella | B. mori | M. sexta | D. plexippus |
|---|---|---|---|---|
| kir1 | g814.t1 * (MT274448) | XM_012695801.2 | XM_030172912 | XM_032669786 |
| kir2 | XM_011554745 (MN894569) | XM_004932424.3 | XM_030172936 | XM_032669772 |
| kir3A | XM_011568587 (MN894570) | XM_004924292.3 | XM_030170470 | XM_032666177 |
| kir3B | XM_011564409 (MN894571) | XM_021346768.1 | XM_030170480 | AGBW02010886 |
| kir4A | XM_011554721 | XM_004932543.3 | XM_030172938 | - |
| kir4B | - | XM_012688607.2 | XM_030182287 | XM_032665754 |
| Channel | PxKir1 (%) | PxKir2 (%) | PxKir3A (%) | PxKir3B (%) | PxKir4 (%) |
|---|---|---|---|---|---|
| PxKir1 | 100 | 50.0 (72.0) | 27.0 (41.7) | 37.7 (45.8) | 36.0 (47.3) |
| PxKir2 | 100 | 32.1 (72.7) | 31.9 (62.1) | 51.9 (59.2) | |
| PxKir3A | 100 | 40.8 (52.4) | 35.3 (38.0) | ||
| PxKir3B | 100 | 34.4 (43.1) | |||
| PxKir4 | 100 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lai, X.; Xu, J.; Ma, H.; Liu, Z.; Zheng, W.; Liu, J.; Zhu, H.; Zhou, Y.; Zhou, X. Identification and Expression of Inward-Rectifying Potassium Channel Subunits in Plutella xylostella. Insects 2020, 11, 461. https://doi.org/10.3390/insects11080461
Lai X, Xu J, Ma H, Liu Z, Zheng W, Liu J, Zhu H, Zhou Y, Zhou X. Identification and Expression of Inward-Rectifying Potassium Channel Subunits in Plutella xylostella. Insects. 2020; 11(8):461. https://doi.org/10.3390/insects11080461
Chicago/Turabian StyleLai, Xiaoyi, Jie Xu, Haihao Ma, Zheming Liu, Wei Zheng, Jia Liu, Hang Zhu, Yong Zhou, and Xiaomao Zhou. 2020. "Identification and Expression of Inward-Rectifying Potassium Channel Subunits in Plutella xylostella" Insects 11, no. 8: 461. https://doi.org/10.3390/insects11080461