(20S) Ginsenoside Rh2 Inhibits STAT3/VEGF Signaling by Targeting Annexin A2
Abstract
:1. Introduction
2. Results
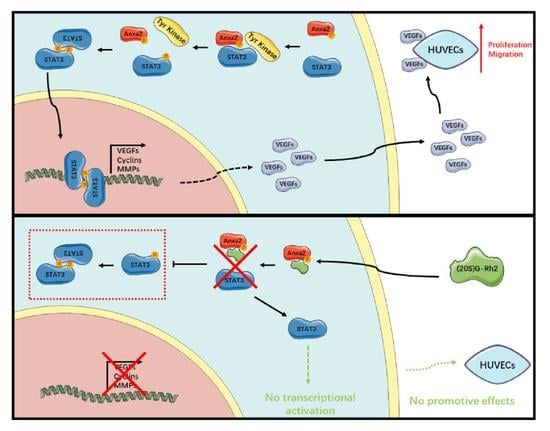
2.1. Anxa2 Regulates VEGF Expression and Secretion
2.2. Anxa2 in Tumor Cells Promotes Proliferation and Migration of Co-Cultured HUVECs
2.3. (20S)G-Rh2 Inhibits STAT3 Activation by Targeting Anxa2
2.4. (20S)G-Rh2 Inhibits Proliferation and Migration of HUVECs
2.5. (20. S)G-Rh2 Interacts with Anxa2-Y23D Mutant
2.6. K301A Mutant of Anxa2 Protects HepG2 Cells from (20S)G-Rh2 Induced STAT3 Inhibition
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Culture
4.2. Chemicals, Antibody, and Plasmids
4.3. Cell Co-Culture Systems
5. Immuno-Precipitation
6. Thermal Shift Assay
7. Dual Luciferase Reporter Assay
8. Real-Time Polymerase Chain Reaction (qRT-PCR)
9. Enzyme-Linked Immune Sorbent Assay (ELISA)
10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Xiong, A.; Yang, Z.; Shen, Y.; Zhou, J.; Shen, Q. Transcription factor STAT3 as a novel molecular target for cancer prevention. Cancers 2014, 16, 926–957. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Lv, J.; Yang, W.; Xu, B.; Wang, Z.; Yu, Z.; Wu, J.; Yang, Y.; Han, Y. Targeted inhibition of STAT3 as a potential treatment strategy for atherosclerosis. Theranostics 2019, 9, 6424–6442. [Google Scholar] [CrossRef] [PubMed]
- Kumari, N.; Dwarakanath, B.S.; Das, A.; Bhatt, A.N. Role of interleukin-6 in cancer progression and therapeutic resistance. Tumour Biol. 2016, 37, 11553–11572. [Google Scholar] [CrossRef] [PubMed]
- Darnell, J.E., Jr.; Kerr, I.M.; Stark, G.R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 1994, 264, 1415–1421. [Google Scholar] [CrossRef] [Green Version]
- Daniel, E.J.; Rachel, A.O.; Jennifer, R.G. Targeting the IL-6 JAK STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 234–248. [Google Scholar] [CrossRef]
- Jennifer, H.; Ashwini, C.; Daniel, G.; Matthias, E. Therapeutically exploiting STAT3 activity in cancer using tissue repair as a road map. Nat. Rev. Cancer 2019, 19, 82–96. [Google Scholar] [CrossRef]
- Gabriella, M.; Tyvette, S.H.; James, T. Therapeutic modulator of STAT signalling for human diseases. Nat. Rev. Drug Discov. 2013, 12, 611–629. [Google Scholar] [CrossRef] [Green Version]
- Eirini, B.; Jacqueline, B. Targeting the tumor microenvironment: JAK-STAT3 signaling. JAKSTAT 2013, 2, e23828. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.; Richard, J. The STATs of cancer—New molecular targets come of age. Nat. Rev. Cancer 2004, 4, 97–105. [Google Scholar] [CrossRef]
- Campbell, G.S.; Meyer, D.J.; Raz, R.; Levy, D.E.; Schwartz, J.; Carter-Su, C. Activation of acute phase response factor (APRF)/Stat3 transcription factor by growth hormone. J. Biol. Chem. 1995, 270, 3974–3979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aliaa, M.M.; Heba, A.H.; Dalia, A.; Mohamed, A.A. STAT3 transcription factor as target for anti-cancer therapy. Pharmacol. Rep. 2020, 72. [Google Scholar] [CrossRef]
- David, A.F. STAT3 as a central mediator of neoplastic cellular transformation. Cancer Lett. 2007, 251, 199–210. [Google Scholar] [CrossRef]
- Roeser, J.C.; Leach, S.D.; McAllister, F. Emerging strategies for cancer immunoprevention. Oncogene 2015, 34, 6029–6039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helene, K.M.; Guo, X.; Li, C.; Lodewijk, V.D. Protein interactions between surface annexin A2 and S100A10 mediate adhesion of breast cancer cells to microvascular endothelial cells. FEBS Lett. 2013, 587, 3210–3215. [Google Scholar] [CrossRef] [Green Version]
- Jyoti, K.; Jesper, N. S100 and annexin proteins identify cell membrane damage as the Achilles heel of metastatic cancer cells. Cell Cycle 2015, 14, 502–509. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Wang, H. Clinical significance of Annexin A2 expression in oral squamous cell carcinoma and its influence on cell proliferation, migration and invasion. Sci. Rep. 2021, 11, 5033. [Google Scholar] [CrossRef]
- Tan, S.H.; Young, D.; Chen, Y.; Kuo, H.C.; Alagarsamy, S.; Albert, D.; Gyorgy, P.; Jennifer, C.; David, G.M.; Inger, L.R.; et al. Prognostic features of Annexin A2 expression in prostate cancer. Pathology 2021, 53, 205–213. [Google Scholar] [CrossRef]
- Mahesh, C.S. Annexin A2 (ANX A2): An emerging biomarker and potential therapeutic target for aggressive cancers. Int. J. Cancer 2019, 144, 2074–2081. [Google Scholar] [CrossRef]
- Murilo, R.R.; Pedro, B.; Annie, C.M.S.; Priscila, V.F.; Ivanir, M.O.; Mariana, B.; Jose, A.M.D. Annexin A2 overexpression associates with colorectal cancer invasiveness and TGF-β induced epithelial mesenchymal transition via Src/ANXA2/STAT3. Sci. Rep. 2018, 8, 11285. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Nie, S.; Lv, Z.; Ma, L.; Song, Y.; Hu, Z.; Hu, X.; Liu, Z.; Zhou, G.; Dai, Z.; et al. Overexpression of Annexin A2 promotes proliferation by forming a Glypican 1/c-Myc positive feedback loop: Prognostic significance in human glioma. Cell Death Dis. 2021, 12, 261. [Google Scholar] [CrossRef]
- Hamdy, S.M.; Kazutaka, K.; Hirohito, Y.; Masaaki, T.; Asahiro, M.; Jian, G.; Shi, L.; Masayuki, M.; Joji, T.; Kiyohito, K.; et al. Annexin A2 expression and phosphorylation are up-regulated in hepatocellular carcinoma. Int. J. Oncol. 2008, 33, 1157–1163. [Google Scholar]
- Jung, H.; Kim, J.S.; Kim, W.K.; Kim, J.M.; Lee, H.J.; Han, B.S.; Kim, D.S.; Seo, Y.S.; Lee, S.C.; Park, S.G.; et al. Intracellular annexin A2 regulates NF-κB signaling by binding to the p50 subunit implications for gemcitabbine resistance in pancreatic. Cell Death Dis. 2015, 6, e1606. [Google Scholar] [CrossRef] [Green Version]
- Yuan, J.; Yang, Y.; Gao, Z.; Wang, Z.; Ji, W.; Song, W.; Zhang, F.; Niu, R. Tyr23 phosphorylation of Anxa2 enhances STAT3 activation and promotes proliferation and invasion of breast cancer cells. Breast Cancer Res. Treat. 2017, 164, 327–340. [Google Scholar] [CrossRef]
- Wang, T.; Yuan, J.; Zhang, J.; Tian, R.; Ji, W.; Zhou, Y.; Yang, Y.; Song, W.; Zhang, F.; Niu, R. Anxa2 binds to STAT3 and promotes epithelial to mesenchymal transition in breast cancer cells. Oncotarget 2015, 6, 30975–30992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Susobhan, D.; Praveenkumar, S.; Mallika, V.; Subhamoy, D.; Zygmunt, G.; Jamboor, K.V. Signal transducer and activator of transcription 6 (STAT6) is a novel interactor of annexin A2 in prostate cancer cells. Biochemistry 2010, 49, 2216–2226. [Google Scholar] [CrossRef]
- Sophia, Z.S.; Yang, P.Y.; Edyta, M.G.; Kayla, N.; Shao, S.; Claudio, Z.; Jonathan, I.; Lara, I.; Caroline, S.; Stormi, R.C.; et al. YAP-dependent proliferation by a small molecule targeting annexin A2. Nat. Chem. Biol. 2021, 17, 767–775. [Google Scholar] [CrossRef]
- Wang, Y.S.; Lin, Y.; Li, Y.; Song, Z.; Jin, Y.H. The identification of molecular target of (20S)Ginsenoside Rh2 for its anti-cancer activity. Sci. Rep. 2017, 7, 12408. [Google Scholar] [CrossRef]
- Wang, Y.S.; Li, H.; Li, Y.; Zhang, S.; Jin, Y.H. (20S)G-Rh2 Inhibits NF-κB Regulated Epithelial-Mesenchymal Transition by Targeting Annexin A2. Biomolecules 2020, 10, 528. [Google Scholar] [CrossRef] [Green Version]
- Rajendra, S.A.; Daniel, S.C.; Napoleone, F. VEGF in Signaling and Disease: Beyond Discovery and Development. Cell 2019, 176, 1248–1264. [Google Scholar] [CrossRef] [Green Version]
- Napoleone, F.; Anthony, P.A. Ten years of anti-vascular endothelial growth factor therapy. Nat. Rev. Drug Discov. 2016, 15, 385–403. [Google Scholar] [CrossRef] [Green Version]
- Kari, A.; Tuomas, T.; Tatiana, V.P. Lymphangiogenesis in development and human disease. Nature 2005, 438, 946–953. [Google Scholar] [CrossRef]
- Napoleone, F. Vascular endothelial growth factor and age-related macular degeneration: From basic science to therapy. Nat. Med. 2010, 16, 1107–1111. [Google Scholar] [CrossRef]
- Michele, D.P.; Daniela, B.; Tatiana, V.P. Microenvironmental regulation of tumour angiogenesis. Nat. Rev. Cancer 2017, 17, 457–474. [Google Scholar] [CrossRef]
- Ann, K.G.; Jaakko, S.; Anni, V. Protein phosphorylation and its role in the regulation of Annexin A2 function. Biochim. Biophys. Acta Gen. Subj. 2017, 1861 Pt A, 2515–2529. [Google Scholar] [CrossRef]
- Natthew, S.W.; Christine, J.W. STAT3 the oncogene—Seluding therapy? FEBS J. 2015, 282, 2600–2611. [Google Scholar] [CrossRef] [Green Version]
- Hemmann, U.; Gerhartz, C.; Heesel, B.; Sasse, J.; Kurapkat, G.; Grotzinger, J.; Wollmer, A.; Zhong, Z.; Darnell, J.E., Jr.; Graeve, L.; et al. Differential activation of acute phase response factor/Stat3 and Stat1 via the cytoplasmic domain of the interleukin 6 signal transducer gp130. II Src homology SH2 domains define the specificity of stat factor activation. J. Biol. Chem. 1996, 271, 12999–13007. [Google Scholar] [CrossRef] [Green Version]
- Lidia, A.; Annalisa, C.; Andrea, C.; Valeria, P. STAT3 in cancer a double-edged sword. Cytokine 2017, 98, 42–50. [Google Scholar] [CrossRef]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.-S.; Chen, C.; Zhang, S.-Y.; Li, Y.; Jin, Y.-H. (20S) Ginsenoside Rh2 Inhibits STAT3/VEGF Signaling by Targeting Annexin A2. Int. J. Mol. Sci. 2021, 22, 9289. https://doi.org/10.3390/ijms22179289
Wang Y-S, Chen C, Zhang S-Y, Li Y, Jin Y-H. (20S) Ginsenoside Rh2 Inhibits STAT3/VEGF Signaling by Targeting Annexin A2. International Journal of Molecular Sciences. 2021; 22(17):9289. https://doi.org/10.3390/ijms22179289
Chicago/Turabian StyleWang, Yu-Shi, Chen Chen, Shi-Yin Zhang, Yang Li, and Ying-Hua Jin. 2021. "(20S) Ginsenoside Rh2 Inhibits STAT3/VEGF Signaling by Targeting Annexin A2" International Journal of Molecular Sciences 22, no. 17: 9289. https://doi.org/10.3390/ijms22179289