Induced Cardiomyocyte Proliferation: A Promising Approach to Cure Heart Failure
Abstract
:1. Introduction
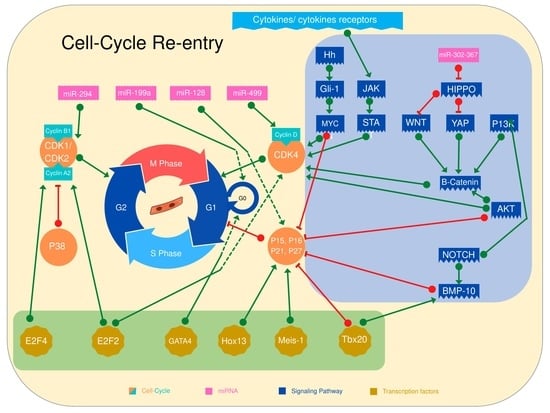
2. Induction of Cardiomyocyte Cell Cycle Entry through Direct Cell Cycle Activation
3. Transcription Factors Control Cardiomyocyte Cell Cycle Re-Entry
4. MicroRNA Regulates Cardiomyocyte Cell Cycle Entry
5. Signaling Pathways That Control Cardiomyocyte Proliferation
6. Metabolic Influence on Cardiomyocyte Division
7. Metabolic Substrates
8. Recent Genetic Tools to Track Cardiomyocyte Cell Cycling
9. The Premise to Cure Heart Failure and Current Obstacles
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Braunwald, E.; Bristow, M.R. Congestive heart failure: Fifty years of progress. Circulation 2000, 102, 14–23. [Google Scholar] [CrossRef]
- Braunwald, E.; Pfeffer, M.A. Ventricular enlargement and remodeling following acute myocardial infarction: Mechanisms and management. Am. J. Cardiol. 1991, 68, 1–6. [Google Scholar] [CrossRef]
- Taylor, D.O.; Edwards, L.B.; Boucek, M.M.; Trulock, E.P.; Deng, M.C.; Keck, B.M.; Hertz, M.I. Registry of the International Society for Heart and Lung Transplantation: Twenty-second Official Adult Heart Transplant Report—2005. J. Hear. Lung Transplant. 2005, 24, 945–955. [Google Scholar] [CrossRef] [PubMed]
- Sturzu, A.C.; Rajarajan, K.; Passer, D.; Plonowska, K.; Riley, A.; Tan, T.C.; Sharma, A.; Xu, A.F.; Engels, M.C.; Feistritzer, R.; et al. Fetal Mammalian Heart Generates a Robust Compensatory Response to Cell Loss. Circulation 2015, 132, 109–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porrello, E.R.; Mahmoud, A.I.; Simpson, E.; Johnson, B.A.; Grinsfelder, D.; Canseco, D.; Mammen, P.P.; Rothermel, B.A.; Olson, E.N.; Sadek, H.A. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc. Natl. Acad. Sci. USA 2013, 110, 187–192. [Google Scholar] [CrossRef] [Green Version]
- Porrello, E.R.; Mahmoud, A.I.; Simpson, E.; Hill, J.A.; Richardson, J.A.; Olson, E.N.; Sadek, H. Transient Regenerative Potential of the Neonatal Mouse Heart. Science 2011, 331, 1078–1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergmann, O.; Zdunek, S.; Felker, A.; Salehpour, M.; Alkass, K.; Bernard, S.; Sjostrom, S.L.; Szewczykowska, M.; Jackowska, T.; Dos Remedios, C.; et al. Dynamics of Cell Generation and Turnover in the Human Heart. Cell 2015, 161, 1566–1575. [Google Scholar] [CrossRef] [Green Version]
- Bergmann, O.; Bhardwaj, R.D.; Bernard, S.; Zdunek, S.; Barnabé-Heider, F.; Walsh, S.; Zupicich, J.; Alkass, K.; Buchholz, B.A.; Druid, H.; et al. Evidence for Cardiomyocyte Renewal in Humans. Science 2009, 324, 98–102. [Google Scholar] [CrossRef] [Green Version]
- Kimura, W.; Xiao, F.; Canseco, D.C.; Muralidhar, S.; Thet, S.; Zhang, H.M.; Abderrahman, Y.; Chen, R.; Garcia, J.A.; Shelton, J.M.; et al. Hypoxia fate mapping identifies cycling cardiomyocytes in the adult heart. Nat. Cell Biol. 2015, 523, 226–230. [Google Scholar] [CrossRef]
- Senyo, S.; Steinhauser, M.L.; Pizzimenti, C.L.; Yang, V.K.; Cai, L.; Wang, M.; Wu, T.-D.; Guerquin-Kern, J.-L.; Lechene, C.P.; Lee, R.T. Mammalian heart renewal by pre-existing cardiomyocytes. Nat. Cell Biol. 2013, 493, 433–436. [Google Scholar] [CrossRef] [Green Version]
- Beltrami, A.P.; Urbanek, K.; Kajstura, J.; Yan, S.; Finato, N.; Bussani, R.; Nadal-Ginard, B.; Silvestri, F.; Leri, A.; Beltrami, C.A.; et al. Evidence That Human Cardiac Myocytes Divide after Myocardial Infarction. N. Engl. J. Med. 2001, 344, 1750–1757. [Google Scholar] [CrossRef]
- Hassink, R.J.; Pasumarthi, K.B.; Nakajima, H.; Rubart, M.; Soonpaa, M.H.; De La Rivière, A.B.; Doevendans, P.A.; Field, L.J. Cardiomyocyte cell cycle activation improves cardiac function after myocardial infarction. Cardiovasc. Res. 2007, 78, 18–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasumarthi, K.B.S.; Nakajima, H.; Nakajima, H.O.; Soonpaa, M.H.; Field, L.J. Targeted Expression of Cyclin D2 Results in Cardiomyocyte DNA Synthesis and Infarct Regression in Transgenic Mice. Circ. Res. 2005, 96, 110–118. [Google Scholar] [CrossRef] [Green Version]
- Soonpaa, M.H.; Field, L.J. Assessment of cardiomyocyte DNA synthesis in normal and injured adult mouse hearts. Am. J. Physiol. Heart Circ. Physiol. 1997, 272, H220–H226. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Meng, Q.; Yu, Y.; Shao, L.; Shen, Z. Adult Cardiomyocyte Proliferation: A New Insight for Myocardial Infarction Therapy. J. Cardiovasc. Transl. Res. 2020. [Google Scholar] [CrossRef]
- Ikenishi, A.; Okayama, H.; Iwamoto, N.; Yoshitome, S.; Tane, S.; Nakamura, K.; Obayashi, T.; Hayashi, T.; Takeuchi, T. Cell cycle regulation in mouse heart during embryonic and postnatal stages. Dev. Growth Differ. 2012, 54, 731–738. [Google Scholar] [CrossRef]
- Kang, M.J. Cyelins and Cyelin Dependent Kinases during Cardiac Development. Mol. Cells 1997, 7, 360–366. [Google Scholar]
- Tane, S.; Ikenishi, A.; Okayama, H.; Iwamoto, N.; Nakayama, K.I.; Takeuchi, T. CDK inhibitors, p21Cip1 and p27Kip1, participate in cell cycle exit of mammalian cardiomyocytes. Biochem. Biophys. Res. Commun. 2014, 443, 1105–1109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tane, S.; Okayama, H.; Ikenishi, A.; Amemiya, Y.; Nakayama, K.I.; Takeuchi, T. Two inhibitory systems and CKIs regulate cell cycle exit of mammalian cardiomyocytes after birth. Biochem. Biophys. Res. Commun. 2015, 466, 147–154. [Google Scholar] [CrossRef]
- Engel, F.; Hauck, L.; Boehm, M.; Nabel, E.G.; Dietz, R.; von Harsdorf, R. p21 CIP1 Controls Proliferating Cell Nuclear Antigen Level in Adult Cardiomyocytes. Mol. Cell. Biol. 2003, 23, 555–565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kochilas, L.K.; Li, J.; Jin, F.; Buck, C.A.; Epstein, J.A. p57Kip2 Expression Is Enhanced During Mid-Cardiac Murine Development and Is Restricted to Trabecular Myocardium. Pediatric Res. 1999, 45, 635–642. [Google Scholar] [CrossRef] [Green Version]
- Aix, E.; Gutiérrez-Gutiérrez, Ó.; Sánchez-Ferrer, C.; Aguado, T.; Flores, I. Postnatal telomere dysfunction induces cardiomyocyte cell-cycle arrest through p21 activation. J. Cell Biol. 2016, 213, 571–583. [Google Scholar] [CrossRef] [Green Version]
- Di Stefano, V.; Giacca, M.; Capogrossi, M.C.; Crescenzi, M.; Martelli, F. Knockdown of Cyclin-dependent Kinase Inhibitors Induces Cardiomyocyte Re-entry in the Cell Cycle. J. Biol. Chem. 2011, 286, 8644–8654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engel, F.B. p38 MAP kinase inhibition enables proliferation of adult mammalian cardiomyocytes. Genes Dev. 2005, 19, 1175–1187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eriksson, M.; Leppä, S. Mitogen-activated Protein Kinases and Activator Protein 1 Are Required for Proliferation and Cardiomyocyte Differentiation of P19 Embryonal Carcinoma Cells. J. Biol. Chem. 2002, 277, 15992–16001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soonpaa, M.H.; Koh, G.Y.; Pajak, L.; Jing, S.; Wang, H.; Franklin, M.T.; Kim, K.K.; Field, L.J. Cyclin D1 overexpression promotes cardiomyocyte DNA synthesis and multinucleation in transgenic mice. J. Clin. Investig. 1997, 99, 2644–2654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Yue, S.; Chen, X.; Kubin, T.; Braun, T. Regulation of cardiomyocyte polyploidy and multinucleation by CyclinG1. Circ. Res. 2010, 106, 1498–1506. [Google Scholar] [CrossRef]
- Tane, S.; Kubota, M.; Okayama, H.; Ikenishi, A.; Yoshitome, S.; Iwamoto, N.; Satoh, Y.; Kusakabe, A.; Ogawa, S.; Kanai, A.; et al. Repression of Cyclin D1 Expression Is Necessary for the Maintenance of Cell Cycle Exit in Adult Mammalian Cardiomyocytes. J. Biol. Chem. 2014, 289, 18033–18044. [Google Scholar] [CrossRef] [Green Version]
- Chaudhry, H.W.; Dashoush, N.H.; Tang, H.; Zhang, L.; Wang, X.; Wu, E.X.; Wolgemuth, D.J. Cyclin A2 Mediates Cardiomyocyte Mitosis in the Postmitotic Myocardium. J. Biol. Chem. 2004, 279, 35858–35866. [Google Scholar] [CrossRef] [Green Version]
- Woo, Y.J. Therapeutic Delivery of Cyclin A2 Induces Myocardial Regeneration and Enhances Cardiac Function in Ischemic Heart Failure. Circulation 2006, 114, I-206–I-213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, R.; Asai, T.; Tang, H.; Dashoush, N.H.; Kara, R.J.; Costa, K.D.; Naka, Y.; Wu, E.X.; Wolgemuth, D.J.; Chaudhry, H.W. Cyclin A2 Induces Cardiac Regeneration After Myocardial Infarction and Prevents Heart Failure. Circ. Res. 2007, 100, 1741–1748. [Google Scholar] [CrossRef] [Green Version]
- Shapiro, S.D.; Ranjan, A.; Kawase, Y.; Cheng, R.; Kara, R.J.; Bhattacharya, R.; Guzmán-Martínez, G.; Sanz, J.; Garcia, M.J.; Chaudhry, H.W. Cyclin A2 Induces Cardiac Regeneration After Myocardial Infarction Through Cytokinesis of Adult Cardiomyocytes. Sci. Transl. Med. 2014, 6, 224ra27. [Google Scholar] [CrossRef]
- Tamamori-Adachi, M.; Ito, H.; Sumrejkanchanakij, P.; Adachi, S.; Hiroe, M.; Shimizu, M.; Kawauchi, J.; Sunamori, M.; Marumo, F.; Kitajima, S.; et al. Critical Role of Cyclin D1 Nuclear Import in Cardiomyocyte Proliferation. Circ. Res. 2003, 92, e12–e19. [Google Scholar] [CrossRef] [Green Version]
- Katrina, C.; Brooks, G. Forced expression of the cyclin B1–CDC2 complex induces proliferation in adult rat cardiomyocytes. Biochem. J. 2004, 382, 411–416. [Google Scholar] [CrossRef]
- Mohamed, T.M.; Ang, Y.-S.; Radzinsky, E.; Zhou, P.; Huang, Y.; Elfenbein, A.; Foley, A.; Magnitsky, S.; Srivastava, D. Regulation of Cell Cycle to Stimulate Adult Cardiomyocyte Proliferation and Cardiac Regeneration. Cell 2018, 173, 104–116.e12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abouleisa, R.; Ou, Q.; Tang, X.; Solanki, M.; Guo, Y.; McNally, L.; Lorkiewicz, P.; Choudhary, K.; Thomas, R.; Juhardeen, H.R.; et al. Transient Cell Cycle Induction in Cardiomyocytes to Treat. Ischemic Heart Failure; Research Square: Durham, NC, USA, 2020. [Google Scholar]
- Li, B.; Li, M.; Li, X.; Li, H.; Lai, Y.; Huang, S.; He, X.; Si, X.; Zheng, H.; Liao, W.; et al. Sirt1-inducible deacetylation of p21 promotes cardiomyocyte proliferation. Aging 2019, 11, 12546–12567. [Google Scholar] [CrossRef]
- Attwooll, C.; Lazzerini Denchi, E.; Helin, K. The E2F family: Specific functions and overlapping interests. EMBO J. 2004, 23, 4709–4716. [Google Scholar] [CrossRef] [PubMed]
- Ebelt, H.; Zhang, Y.; Kampke, A.; Xu, J.; Schlitt, A.; Buerke, M.; Müller-Werdan, U.; Werdan, K.; Braun, T. E2F2 expression induces proliferation of terminally differentiated cardiomyocytes in vivo. Cardiovasc. Res. 2008, 80, 219–226. [Google Scholar] [CrossRef] [PubMed]
- van Amerongen, M.J.; Diehl, F.; Novoyatleva, T.; Patra, C.; Engel, F.B. E2F4 is required for cardiomyocyte proliferation. Cardiovasc. Res. 2010, 86, 92–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sdek, P.; Zhao, P.; Wang, Y.; Huang, C.-J.; Ko, C.Y.; Butler, P.; Weiss, J.N.; MacLellan, W.R. Rb and p130 control cell cycle gene silencing to maintain the postmitotic phenotype in cardiac myocytes. J. Cell Biol. 2011, 194, 407–423. [Google Scholar] [CrossRef] [Green Version]
- MacLellan, W.R.; Garcia, A.; Oh, H.; Frenkel, P.; Jordan, M.C.; Roos, K.P.; Schneider, M. Overlapping Roles of Pocket Proteins in the Myocardium Are Unmasked by Germ Line Deletion of p130 plus Heart-Specific Deletion of Rb. Mol. Cell. Biol. 2005, 25, 2486–2497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agah, R.; A Kirshenbaum, L.; Abdellatif, M.; Truong, L.D.; Chakraborty, S.; Michael, L.H.; Schneider, M.D. Adenoviral delivery of E2F-1 directs cell cycle reentry and p53-independent apoptosis in postmitotic adult myocardium in vivo. J. Clin. Investig. 1997, 100, 2722–2728. [Google Scholar] [CrossRef] [PubMed]
- Yurkova, N.; Shaw, J.; Blackie, K.; Weidman, D.; Jayas, R.; Flynn, B.; Kirshenbaum, L.A. The Cell Cycle Factor E2F-1 Activates Bnip3 and the Intrinsic Death Pathway in Ventricular Myocytes. Circ. Res. 2008, 102, 472–479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azcoitia, V.; Aracil, M.; Martinez, A.C.; Torres, M. The homeodomain protein Meis1 is essential for definitive hematopoiesis and vascular patterning in the mouse embryo. Dev. Biol. 2005, 280, 307–320. [Google Scholar] [CrossRef] [Green Version]
- Jopling, C.; Sleep, E.; Raya, M.; Martí, M.; Raya, A.; Belmonte, J.C.I. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 2010, 464, 606–609. [Google Scholar] [CrossRef]
- Hisa, T.; E Spence, S.; A Rachel, R.; Fujita, M.; Nakamura, T.; Ward, J.M.; E Devor-Henneman, D.; Saiki, Y.; Kutsuna, H.; Tessarollo, L.; et al. Hematopoietic, angiogenic and eye defects in Meis1 mutant animals. EMBO J. 2004, 23, 450–459. [Google Scholar] [CrossRef] [Green Version]
- Imamura, T.; Morimoto, A.; Takanashi, M.; Hibi, S.; Sugimoto, T.; Ishii, E.; Imashuku, S. Frequent co-expression of HoxA9 and Meis1 genes in infant acute lymphoblastic leukaemia with MLL rearrangement. Br. J. Haematol. 2002, 119, 119–121. [Google Scholar] [CrossRef]
- Paige, S.L.; Thomas, S.; Stoick-Cooper, C.L.; Wang, H.; Maves, L.; Sandstrom, R.; Pabon, L.; Reinecke, H.; Pratt, G.; Keller, G.; et al. A Temporal Chromatin Signature in Human Embryonic Stem Cells Identifies Regulators of Cardiac Development. Cell 2012, 151, 221–232. [Google Scholar] [CrossRef] [Green Version]
- Wamstad, J.A.; Alexander, J.; Truty, R.M.; Shrikumar, A.; Li, F.; Eilertson, K.; Ding, H.; Wylie, J.N.; Pico, A.; Capra, J.A.; et al. Dynamic and Coordinated Epigenetic Regulation of Developmental Transitions in the Cardiac Lineage. Cell 2012, 151, 206–220. [Google Scholar] [CrossRef] [Green Version]
- Mahmoud, A.I.; Kocabas, F.; Muralidhar, S.A.; Kimura, W.; Koura, A.S.; Thet, S.; Porrello, E.; Sadek, H.A. Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nat. Cell Biol. 2013, 497, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Showell, C.; Binder, O.; Conlon, F.L. T-box genes in early embryogenesis. Dev. Dyn. 2004, 229, 201–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greulich, F.; Rudat, C.; Kispert, A. Mechanisms of T-box gene function in the developing heart. Cardiovasc. Res. 2011, 91, 212–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiang, F.L.; Guo, M.; Yutzey, K.E. Overexpression of Tbx20 in Adult Cardiomyocytes Promotes Proliferation and Improves Cardiac Function After Myocardial Infarction. Circulation 2016, 133, 1081–1092. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, S.; Sengupta, A.; Yutzey, K.E. Tbx20 promotes cardiomyocyte proliferation and persistence of fetal characteristics in adult mouse hearts. J. Mol. Cell Cardiol. 2013, 62, 203–213. [Google Scholar] [CrossRef]
- Shen, T.; Aneas, I.; Sakabe, N.; Dirschinger, R.J.; Wang, G.; Smemo, S.; Westlund, J.M.; Cheng, H.; Dalton, N.; Gu, Y.; et al. Tbx20 regulates a genetic program essential to adult mouse cardiomyocyte function. J. Clin. Investig. 2011, 121, 4640–4654. [Google Scholar] [CrossRef] [Green Version]
- Monzen, K.; Shiojima, I.; Hiroi, Y.; Kudoh, S.; Oka, T.; Takimoto, E.; Hayashi, D.; Hosoda, T.; Habara-Ohkubo, A.; Nakaoka, T.; et al. Bone Morphogenetic Proteins Induce Cardiomyocyte Differentiation through the Mitogen-Activated Protein Kinase Kinase Kinase TAK1 and Cardiac Transcription Factors Csx/Nkx-2.5 and GATA-4. Mol. Cell. Biol. 1999, 19, 7096–7105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molkentin, J.D.; Lin, Q.; Duncan, S.A.; Olson, E.N. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997, 11, 1061–1072. [Google Scholar] [CrossRef] [Green Version]
- Mohammadi, M.M.; Kattih, B.; Grund, A.; Froese, N.; Korf-Klingebiel, M.; Gigina, A.; Schrameck, U.; Rudat, C.; Liang, Q.; Kispert, A.; et al. The transcription factor GATA 4 promotes myocardial regeneration in neonatal mice. EMBO Mol. Med. 2017, 9, 265–279. [Google Scholar] [CrossRef]
- Nguyen, N.U.N.; Canseco, D.C.; Xiao, F.; Nakada, Y.; Li, S.; Lam, N.T.; Muralidhar, S.A.; Savla, J.J.; Hill, J.A.; Le, V.; et al. A calcineurin–Hoxb13 axis regulates growth mode of mammalian cardiomyocytes. Nat. Cell Biol. 2020, 582, 271–276. [Google Scholar] [CrossRef]
- Barwari, T.; Joshi, A.; Mayr, M. MicroRNAs in Cardiovascular Disease. J. Am. Coll. Cardiol. 2016, 68, 2577–2584. [Google Scholar] [CrossRef] [Green Version]
- Tian, Y.; Liu, Y.; Wang, T.; Zhou, N.; Kong, J.; Chen, L.; Snitow, M.; Morley, M.; Melinda, S.; Petrenko, N.; et al. A microRNA-Hippo pathway that promotes cardiomyocyte proliferation and cardiac regeneration in mice. Sci. Transl. Med. 2015, 7, 279ra38. [Google Scholar] [CrossRef] [Green Version]
- Eulalio, A.; Mano, M.; Ferro, M.D.; Zentilin, L.; Sinagra, G.; Zacchigna, S.; Giacca, M. Functional screening identifies miRNAs inducing cardiac regeneration. Nat. Cell Biol. 2012, 492, 376–381. [Google Scholar] [CrossRef]
- Gabisonia, K.; Prosdocimo, G.; Aquaro, G.D.; Carlucci, L.; Zentilin, L.; Secco, I.; Ali, H.; Braga, L.; Gorgodze, N.; Bernini, F.; et al. MicroRNA therapy stimulates uncontrolled cardiac repair after myocardial infarction in pigs. Nature 2019, 569, 418–422. [Google Scholar] [CrossRef]
- Huang, W.; Feng, Y.; Liang, J.; Yu, H.; Wang, C.; Wang, B.; Wang, M.; Jiang, L.; Meng, W.; Cai, W.; et al. Loss of microRNA-128 promotes cardiomyocyte proliferation and heart regeneration. Nat. Commun. 2018, 9, 700. [Google Scholar] [CrossRef] [Green Version]
- Borden, A.; Kurian, J.; Nickoloff, E.; Yang, Y.; Troupes, C.D.; Ibetti, J.; Lucchese, A.M.; Gao, E.; Mohsin, S.; Koch, W.J.; et al. Transient Introduction of miR-294 in the Heart Promotes Cardiomyocyte Cell Cycle Reentry After Injury. Circ. Res. 2019, 125, 14–25. [Google Scholar] [CrossRef]
- Li, X.; Wang, J.; Jia, Z.; Cui, Q.; Zhang, C.; Wang, W.; Chen, P.; Ma, K.; Zhou, C. MiR-499 Regulates Cell Proliferation and Apoptosis during Late-Stage Cardiac Differentiation via Sox6 and Cyclin D1. PLoS ONE 2013, 8, e74504. [Google Scholar] [CrossRef]
- Barroso-Deljesus, A.; Romero-López, C.; Lucena-Aguilar, G.; Melen, G.; Sanchez, L.; Ligero, G.; Berzal-Herranz, A.; Menendez, P. Embryonic Stem Cell-Specific miR302-367 Cluster: Human Gene Structure and Functional Characterization of Its Core Promoter. Mol. Cell. Biol. 2008, 28, 6609–6619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ambrosy, A.P.; Fonarow, G.; Butler, J.; Chioncel, O.; Greene, S.J.; Vaduganathan, M.; Nodari, S.; Lam, C.S.; Sato, N.; Shah, A.N.; et al. The Global Health and Economic Burden of Hospitalizations for Heart Failure. J. Am. Coll. Cardiol. 2014, 63, 1123–1133. [Google Scholar] [CrossRef] [PubMed]
- Hashmi, S.; Ahmad, H.R. Molecular switch model for cardiomyocyte proliferation. Cell Regen. 2019, 8, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Brazil, D.P.; Yang, Z.-Z.; Hemmings, B.A. Advances in protein kinase B signalling: AKTion on multiple fronts. Trends Biochem. Sci. 2004, 29, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Maddika, S.; Ande, S.R.; Wiechec, E.; Hansen, L.L.; Wesselborg, S.; Los, M. Akt-mediated phosphorylation of CDK2 regulates its dual role in cell cycle progression and apoptosis. J. Cell Sci. 2008, 121, 979–988. [Google Scholar] [CrossRef] [Green Version]
- Parekh, P.; Motiwale, L.; Naik, N.; Rao, K.V.K. Downregulation of cyclin D1 is associated with decreased levels of p38 MAP kinases, Akt/PKB and Pak1 during chemopreventive effects of resveratrol in liver cancer cells. Exp. Toxicol. Pathol. 2011, 63, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Beigi, F.; Schmeckpeper, J.; Pow-Anpongkul, P.; Payne, J.A.; Zhang, L.; Zhang, Z.; Huang, J.; Mirotsou, M.; Dzau, V.J. C3orf58, a Novel Paracrine Protein, Stimulates Cardiomyocyte Cell-Cycle Progression Through the PI3K–AKT–CDK7 Pathway. Circ. Res. 2013, 113, 372–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Z.; Zhou, P.; Von Gise, A.; Gu, F.; Ma, Q.; Chen, J.; Guo, H.; Van Gorp, P.R.; Wang, D.-Z.; Pu, W.T. Pi3kcbLinks Hippo-YAP and PI3K-AKT Signaling Pathways to Promote Cardiomyocyte Proliferation and Survival. Circ. Res. 2015, 116, 35–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gude, N.; Muraski, J.; Rubio, M.; Kajstura, J.; Schaefer, E.; Anversa, P.; Sussman, M.A. Akt Promotes Increased Cardiomyocyte Cycling and Expansion of the Cardiac Progenitor Cell Population. Circ. Res. 2006, 99, 381–388. [Google Scholar] [CrossRef] [Green Version]
- Sussman, M.A.; Völkers, M.; Fischer, K.; Bailey, B.; Cottage, C.T.; Din, S.; Gude, N.; Avitabile, D.; Alvarez, R.; Sundararaman, B.; et al. Myocardial AKT: The Omnipresent Nexus. Physiol. Rev. 2011, 91, 1023–1070. [Google Scholar] [CrossRef]
- von Gise, A.; Lin, Z.; Schlegelmilch, K.; Honor, L.; Pan, G.M.; Buck, J.N.; Ma, Q.; Ishiwata, T.; Zhou, B.; Camargo, F.D.; et al. YAP1, the nuclear target of Hippo signaling, stimulates heart growth through cardiomyocyte proliferation but not hypertrophy. Proc. Natl. Acad. Sci. USA 2012, 109, 2394–2399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xin, M.; Kim, Y.; Sutherland, L.B.; Qi, X.; McAnally, J.; Schwartz, R.J.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. Regulation of Insulin-Like Growth Factor Signaling by Yap Governs Cardiomyocyte Proliferation and Embryonic Heart Size. Sci. Signal. 2011, 4, ra70. [Google Scholar] [CrossRef] [Green Version]
- Gründl, M.; Walz, S.; Hauf, L.; Schwab, M.; Werner, K.M.; Spahr, S.; Schulte, C.; Maric, H.M.; Ade, C.P.; Gaubatz, S. Interaction of YAP with the Myb-MuvB (MMB) complex defines a transcriptional program to promote the proliferation of cardiomyocytes. PLoS Genet. 2020, 16, e1008818. [Google Scholar] [CrossRef]
- Monroe, T.; Hill, M.; Morikawa, Y.; Leach, J.; Heallen, T.; Cao, S.; Krijger, P.; de Laat, W.; Wehrens, X.; Rodney, G.; et al. YAP Partially Reprograms Chromatin Accessibility to Directly Induce Adult Cardiogenesis In Vivo. Dev. Cell 2019, 48, 765–779.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xin, M.; Kim, Y.; Sutherland, L.B.; Murakami, M.; Qi, X.; McAnally, J.; Porrello, E.; Mahmoud, A.I.; Tan, W.; Shelton, J.M.; et al. Hippo pathway effector Yap promotes cardiac regeneration. Proc. Natl. Acad. Sci. USA 2013, 110, 13839–13844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Z.; Von Gise, A.; Zhou, P.; Gu, F.; Ma, Q.; Jiang, J.; Yau, A.L.; Buck, J.N.; Gouin, K.; Van Gorp, P.R.; et al. Cardiac-Specific YAP Activation Improves Cardiac Function and Survival in an Experimental Murine MI Model. Circ. Res. 2014, 115, 354–363. [Google Scholar] [CrossRef] [PubMed]
- Grego-Bessa, J.; Zurita, L.L.; del Monte, G.; Bolós, V.; Melgar, P.; Arandilla, A.; Garratt, A.; Zang, H.; Mukouyama, Y.-S.; Chen, H.; et al. Notch Signaling Is Essential for Ventricular Chamber Development. Dev. Cell 2007, 12, 415–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Shi, S.; Acosta, L.; Li, W.; Lu, J.; Bao, S.; Chen, Z.; Yang, Z.; Schneider, M.D.; Chien, K.R.; et al. BMP10 is essential for maintaining cardiac growth during murine cardiogenesis. Development 2004, 131, 2219–2231. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Borikova, A.L.; Ben-Yair, R.; Guner-Ataman, B.; MacRae, C.A.; Lee, R.T.; Burns, C.G. Notch signaling regulates cardiomyocyte proliferation during zebrafish heart regeneration. Proc. Natl. Acad. Sci. USA 2014, 111, 1403–1408. [Google Scholar] [CrossRef] [Green Version]
- Gude, N.A.; Emmanuel, G.; Wu, W.; Cottage, C.T.; Fischer, K.; Quijada, P.; Muraski, J.A.; Alvarez, R.; Rubio, M.; Schaefer, E.; et al. Activation of Notch-Mediated Protective Signaling in the Myocardium. Circ. Res. 2008, 102, 1025–1035. [Google Scholar] [CrossRef]
- Bergmann, M.W. WNT Signaling in Adult Cardiac Hypertrophy and Remodeling. Circ. Res. 2010, 107, 1198–1208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, K.; Luo, M.; Zhang, Y.; Wilkes, D.C.; Ge, G.; Grieskamp, T.; Yamada, C.; Liu, T.-C.; Huang, G.; Basson, C.T.; et al. Secreted Frizzled-related protein 2 is a procollagen C proteinase enhancer with a role in fibrosis associated with myocardial infarction. Nat. Cell Biol. 2008, 11, 46–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tseng, A.-S.; Engel, F.B.; Keating, M.T. The GSK-3 Inhibitor BIO Promotes Proliferation in Mammalian Cardiomyocytes. Chem. Biol. 2006, 13, 957–963. [Google Scholar] [CrossRef] [Green Version]
- Kerkela, R.; Kockeritz, L.; Macaulay, K.; Zhou, J.; Doble, B.; Beahm, C.; Greytak, S.; Woulfe, K.; Trivedi, C.; Woodgett, J.; et al. Deletion of GSK-3β in mice leads to hypertrophic cardiomyopathy secondary to cardiomyoblast hyperproliferation. J. Clin. Investig. 2008, 118, 3609–3618. [Google Scholar] [CrossRef] [Green Version]
- Fang, Y.; Gupta, V.; Karra, R.; Holdway, J.E.; Kikuchi, K.; Poss, K.D. Translational profiling of cardiomyocytes identifies an early Jak1/Stat3 injury response required for zebrafish heart regeneration. Proc. Natl. Acad. Sci. USA 2013, 110, 13416–13421. [Google Scholar] [CrossRef] [Green Version]
- Miyawaki, A.; Obana, M.; Mitsuhara, Y.; Orimoto, A.; Nakayasu, Y.; Yamashita, T.; Fukada, S.-I.; Maeda, M.; Nakayama, H.; Fujio, Y. Adult murine cardiomyocytes exhibit regenerative activity with cell cycle reentry through STAT3 in the healing process of myocarditis. Sci. Rep. 2017, 7, 1407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunaeva, M.; Waltenberger, J. Hh signaling in regeneration of the ischemic heart. Cell. Mol. Life Sci. 2017, 74, 3481–3490. [Google Scholar] [CrossRef]
- Dyer, L.A.; Kirby, M.L. Sonic hedgehog maintains proliferation in secondary heart field progenitors and is required for normal arterial pole formation. Dev. Biol. 2009, 330, 305–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, W.-Y.; Gemberling, M.; Wang, J.; Holdway, J.E.; Shen, M.-C.; Karlstrom, R.O.; Poss, K.D. In vivo monitoring of cardiomyocyte proliferation to identify chemical modifiers of heart regeneration. Development 2013, 140, 660–666. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Cao, J.; Dickson, A.L.; Poss, K.D. Epicardial regeneration is guided by cardiac outflow tract and Hedgehog signalling. Nature 2015, 522, 226–230. [Google Scholar] [CrossRef] [Green Version]
- Singh, B.N.; Kren, S.M.; Gong, W.; Weaver, C.; Bowlin, K.; Braunlin, E.; Garry, M.G.; Koyano-Nakagawa, N.; Garry, D.J. Abstract 20451: Hedgehog Signaling and Cardiomyocyte Proliferation. Circulation 2014, 130, 20451. [Google Scholar] [CrossRef]
- Kusano, K.F.; Pola, R.; Murayama, T.; Curry, C.; Kawamoto, A.; Iwakura, A.; Shintani, S.; Ii, M.; Asai, J.; Tkebuchava, T.; et al. Sonic hedgehog myocardial gene therapy: Tissue repair through transient reconstitution of embryonic signaling. Nat. Med. 2005, 11, 1197–1204. [Google Scholar] [CrossRef]
- Johnson, N.R.; Wang, Y. Controlled Delivery of Sonic Hedgehog Morphogen and Its Potential for Cardiac Repair. PLoS ONE 2013, 8, e63075. [Google Scholar] [CrossRef] [Green Version]
- Paulis, L.; Fauconnier, J.; Cazorla, O.; Thireau, J.; Soleti, R.; Vidal, B.; Ouillé, A.; Bartholome, M.; Bideaux, P.; Roubille, F.; et al. Activation of Sonic hedgehog signaling in ventricular cardiomyocytes exerts cardioprotection against ischemia reperfusion injuries. Sci. Rep. 2015, 5, 7983. [Google Scholar] [CrossRef] [Green Version]
- Singh, B.; Koyano-Nakagawa, N.; Gong, W.; Moskowitz, I.; Weaver, C.V.; Braunlin, E.; Das, S.; Van Berlo, J.H.; Garry, M.G.; Garry, D.J. A conserved HH-Gli1-Mycn network regulates heart regeneration from newt to human. Nat. Commun. 2018, 9, 4237. [Google Scholar] [CrossRef]
- White, I.A.; Gordon, J.; Balkan, W.; Hare, J.M. Sympathetic Reinnervation Is Required for Mammalian Cardiac Regeneration. Circ. Res. 2015, 117, 990–994. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Zhang, C.-H.; Ammanamanchi, N.; Suresh, S.; Lewarchik, C.; Rao, K.; Uys, G.M.; Han, L.; Abrial, M.; Yimlamai, D.; et al. Control of cytokinesis by β-adrenergic receptors indicates an approach for regulating cardiomyocyte endowment. Sci. Transl. Med. 2019, 11, eaaw6419. [Google Scholar] [CrossRef]
- Mahmoud, A.I.; O’Meara, C.C.; Gemberling, M.; Zhao, L.; Bryant, D.M.; Zheng, R.; Gannon, J.B.; Cai, L.; Choi, W.-Y.; Egnaczyk, G.F.; et al. Nerves Regulate Cardiomyocyte Proliferation and Heart Regeneration. Dev. Cell 2015, 34, 387–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakada, Y.; Canseco, D.C.; Thet, S.; Abdisalaam, S.; Asaithamby, S.A.A.; Santos, C.S.X.D.C.D.; Shah, C.X.S.A.; Zhang, H.; Faber, H.Z.J.E.; Kinter, M.; et al. Hypoxia induces heart regeneration in adult mice. Nat. Cell Biol. 2017, 541, 222–227. [Google Scholar] [CrossRef]
- Guimarães-Camboa, N.; Stowe, J.; Aneas, I.; Sakabe, N.; Cattaneo, P.; Henderson, L.; Kilberg, M.S.; Johnson, R.; Chen, J.; McCulloch, A.D.; et al. HIF1α Represses Cell Stress Pathways to Allow Proliferation of Hypoxic Fetal Cardiomyocytes. Dev. Cell 2015, 33, 507–521. [Google Scholar] [CrossRef] [Green Version]
- Lopaschuk, G.D.; Jaswal, J.S. Energy metabolic phenotype of the cardiomyocyte during development, differentiation, and postnatal maturation. J. Cardiovasc. Pharmacol. 2010, 56, 130–140. [Google Scholar] [CrossRef]
- Cardoso, A.C.; Lam, N.T.; Savla, J.J.; Nakada, Y.; Pereira, A.H.M.; Elnwasany, A.; Menendez-Montes, I.; Ensley, E.L.; Petric, U.B.; Sharma, G.; et al. Mitochondrial substrate utilization regulates cardiomyocyte cell-cycle progression. Nat. Metab. 2020, 2, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Magadum, A.; Singh, N.; Kurian, A.A.; Munir, I.; Mehmood, T.; Brown, K.; Sharkar, M.T.K.; Chepurko, E.; Sassi, Y.; Oh, J.G.; et al. Pkm2 Regulates Cardiomyocyte Cell Cycle and Promotes Cardiac Regeneration. Circulation 2020, 141, 1249–1265. [Google Scholar] [CrossRef] [PubMed]
- Hauck, L.; Dadson, K.; Chauhan, S.; Grothe, D.; Billia, F. Inhibiting the Pkm2/b-catenin axis drives in vivo replication of adult cardiomyocytes following experimental MI. Cell Death Differ. 2020. [Google Scholar] [CrossRef]
- D’Uva, G.; Tzahor, E. The key roles of ERBB2 in cardiac regeneration. Cell Cycle 2015, 14, 2383–2384. [Google Scholar] [CrossRef] [Green Version]
- Honkoop, H.; De Bakker, D.E.; Aharonov, A.; Kruse, F.; Shakked, A.; Nguyen, P.; De Heus, C.; Garric, L.; Muraro, M.J.; Shoffner, A.; et al. Single-cell analysis uncovers that metabolic reprogramming by ErbB2 signaling is essential for cardiomyocyte proliferation in the regenerating heart. eLife 2019, 8, 8. [Google Scholar] [CrossRef]
- Bae, J.; Salamon, R.J.; Brandt, E.B.; Paltzer, W.G.; Zhang, Z.; Britt, E.C.; Hacker, T.A.; Fan, J.; Mahmoud, A.I. Malonate Promotes Adult Cardiomyocyte Proliferation and Heart Regeneration. Circulation 2021, 143, 1973–1986. [Google Scholar] [CrossRef] [PubMed]
- Mills, R.J.; Parker, B.L.; Quaife-Ryan, G.; Voges, H.K.; Needham, E.; Bornot, A.; Ding, M.; Andersson, H.; Polla, M.; Elliott, D.A.; et al. Drug Screening in Human PSC-Cardiac Organoids Identifies Pro-proliferative Compounds Acting via the Mevalonate Pathway. Cell Stem Cell 2019, 24, 895–907.e6. [Google Scholar] [CrossRef] [PubMed]
- Buhaescu, I.; Izzedine, H. Mevalonate pathway: A review of clinical and therapeutical implications. Clin. Biochem. 2007, 40, 575–584. [Google Scholar] [CrossRef] [PubMed]
- Derks, W.; Bergmann, O. Polyploidy in Cardiomyocytes: Roadblock to Heart Regeneration? Circ. Res. 2020, 126, 552–565. [Google Scholar] [CrossRef] [PubMed]
- Leone, M.; Musa, G.; Engel, F.B. Cardiomyocyte binucleation is associated with aberrant mitotic microtubule distribution, mislocalization of RhoA and IQGAP3, as well as defective actomyosin ring anchorage and cleavage furrow ingression. Cardiovasc. Res. 2018, 114, 1115–1131. [Google Scholar] [CrossRef] [PubMed]
- Leone, M.; Engel, F.B. Pseudo-bipolar spindle formation and cell division in postnatal binucleated cardiomyocytes. J. Mol. Cell Cardiol. 2019, 134, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Leone, M.; Engel, F.B. Advances in heart regeneration based on cardiomyocyte proliferation and regenerative potential of binucleated cardiomyocytes and polyploidization. Clin. Sci. 2019, 133, 1229–1253. [Google Scholar] [CrossRef] [PubMed]
- Zong, H.; Espinosa, J.S.; Su, H.H.; Muzumdar, M.D.; Luo, L. Mosaic analysis with double markers in mice. Cell 2005, 121, 479–492. [Google Scholar] [CrossRef] [Green Version]
- Ali, S.R.; Hippenmeyer, S.; Saadat, L.V.; Luo, L.; Weissman, I.L.; Ardehali, R. Existing cardiomyocytes generate cardiomyocytes at a low rate after birth in mice. Proc. Natl. Acad. Sci. USA 2014, 111, 8850–8855. [Google Scholar] [CrossRef] [Green Version]
- Hesse, M.; Doengi, M.; Becker, A.; Kimura, K.; Voeltz, N.; Stein, V.; Fleischmann, B.K. Midbody Positioning and Distance Between Daughter Nuclei Enable Unequivocal Identification of Cardiomyocyte Cell Division in Mice. Circ. Res. 2018, 123, 1039–1052. [Google Scholar] [CrossRef] [PubMed]
- Sakaue-Sawano, A.; Kurokawa, H.; Morimura, T.; Hanyu, A.; Hama, H.; Osawa, H.; Kashiwagi, S.; Fukami, K.; Miyata, T.; Miyoshi, H.; et al. Visualizing Spatiotemporal Dynamics of Multicellular Cell-Cycle Progression. Cell 2008, 132, 487–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magadum, A.; Ding, Y.; He, L.; Kim, T.; Vasudevarao, M.D.; Long, Q.; Yang, K.; Wickramasinghe, N.; Renikunta, H.V.; Dubois, N.; et al. Live cell screening platform identifies PPARδ as a regulator of cardiomyocyte proliferation and cardiac repair. Cell Res. 2017, 27, 1002–1019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jr, R.A.; Wang, B.J.; Quijada, P.J.; Avitabile, D.; Ho, T.; Shaitrit, M.; Chavarria, M.; Firouzi, F.; Ebeid, D.; Monsanto, M.M.; et al. Cardiomyocyte cell cycle dynamics and proliferation revealed through cardiac-specific transgenesis of fluorescent ubiquitinated cell cycle indicator (FUCCI). J. Mol. Cell. Cardiol. 2019, 127, 154–164. [Google Scholar] [CrossRef]
- Uribe, V.; Ramadass, R.; Dogra, D.; Rasouli, S.J.; Gunawan, F.; Nakajima, H.; Chiba, A.; Reischauer, S.; Mochizuki, N.; Stainier, D.Y.R. In vivo analysis of cardiomyocyte proliferation during trabeculation. Development 2018, 145, dev164194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, Y.; Hellwarth, P.B.; Randolph, L.N.; Sun, Y.; Xing, Y.; Zhu, W.; Lian, X.L.; Bao, X. Fluorescent indicators for continuous and lineage-specific reporting of cell-cycle phases in human pluripotent stem cells. Biotechnol. Bioeng. 2020, 117, 2177–2186. [Google Scholar] [CrossRef] [PubMed]
- Bradley, L.A.; Young, A.; Li, H.; Billcheck, H.O.; Wolf, M.J. Loss of Endogenously Cycling Adult Cardiomyocytes Worsens Myocardial Function. Circ. Res. 2021, 128, 155–168. [Google Scholar] [CrossRef]
- Wang, W.E.; Li, L.; Xia, X.; Fu, W.; Liao, Q.; Lan, C.; Yang, D.; Chen, H.; Yue, R.; Zeng, C.S.; et al. Dedifferentiation, Proliferation, and Redifferentiation of Adult Mammalian Cardiomyocytes After Ischemic Injury. Circulation 2017, 136, 834–848. [Google Scholar] [CrossRef]
- Heinzel, F.R.; Hohendanner, F.; Jin, G.; Sedej, S.; Edelmann, F. Myocardial hypertrophy and its role in heart failure with preserved ejection fraction. J. Appl. Physiol. 2015, 119, 1233–1242. [Google Scholar] [CrossRef]
| Cell Cycle Gene | Studied Species | Highlights | References |
|---|---|---|---|
| Cyclin D1 | Adult transgenic mice |
| [26,28] |
| Cyclin D2 | Adult transgenic mice |
| [13] |
| Cyclin G1 | Mice hearts |
| [27] |
| Cyclin A2 | Transgenic mice Rats, pigs |
| [29,30,31,32] |
| CDK4–cyclin D1 | Sprague–Dawley rats |
| [33] |
| Cyclin B1–CDC2AF | Fetal, neonatal, and adult cardiomyocytes isolated from Wistar rat cardiomyocytes |
| [34] |
| CDK1, CDK4, cyclin B1, and cyclin D1 | Post-mitotic mouse, rat, and human cardiomyocytes |
| [35,36] |
| P21 deacetylation by Sirt-1 | Mice hearts |
| [37] |
| Cyclin-dependent kinase inhibitors (CKIs): P21, P27, and P57 | SWISS CD1 mice Wistar rats |
| [23] |
| p38 MAP kinase inhibition | Rat cardiomyocytes |
| [24] |
| Transcription Factor | Studied Species | Highlights | References |
|---|---|---|---|
| E2F | siRNA-mediated knockdown in neonatal mouse and rat cardiomyocytes |
| [39,40] |
| Adenovirus expression in mouse heart |
| ||
| Meis1 | Knockout mice |
| [51] |
| Tbx20 | Induced specific overexpression in an adult mouse |
| [54] |
| Gata4 |
|
| [59] |
| HOX13 | Knockout mice |
| [60] |
| miRNA | Studied Species | Highlights | References |
|---|---|---|---|
| miR-302-367 | Transgenic mice |
| [68] |
| miR-499 | P19CL6 cells |
| [67] |
| miR-199a | AAV overexpressing mirRNA199a |
| [64,69] |
| miR-128 | KO mice |
| [65] |
| miR-294 | AAV overexpression |
| [66] |
| Pathway | Studied Species | Highlights | References |
|---|---|---|---|
| PI3K-AKT | Isolated rat cardiomyocytes |
| [74] |
| Hippo signaling pathway | Neonatal and adult mice cardiomyocytes. Transgenic mice |
| [81,82,83] |
| Notch signaling pathway | Zebrafish |
| [86] |
| Wnt/β-catenin | sFRP2 and GSK3 β knockout mice |
| [89,90,91] |
| Jak/Stat | Zebrafish Adult mice cardiomyocytes |
| [92,93] |
| Hedgehog (Hh) | Zebrafish Newts |
| [102] |
| Autonomic innervation | Zebrafish Mice Human pediatric patients with F4 |
| [103,104,105] |
| Metabolic Factor | Gene Involved | Highlights | Reference |
|---|---|---|---|
| Hypoxia | HIF-alpha |
| [106,107] |
| Mitochondrial substrate utilization | PDK4 |
| [109] |
| Glycolysis | Pkm2 |
| [110,111] |
| Switch from oxidative phosphorylation to glycolysis and lactate fermentation | ErB2 |
| [112,113] |
| Succinate | SDH |
| [114] |
| Mevalonate anabolic pathway |
| [116] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salama, A.B.M.; Gebreil, A.; Mohamed, T.M.A.; Abouleisa, R.R.E. Induced Cardiomyocyte Proliferation: A Promising Approach to Cure Heart Failure. Int. J. Mol. Sci. 2021, 22, 7720. https://doi.org/10.3390/ijms22147720
Salama ABM, Gebreil A, Mohamed TMA, Abouleisa RRE. Induced Cardiomyocyte Proliferation: A Promising Approach to Cure Heart Failure. International Journal of Molecular Sciences. 2021; 22(14):7720. https://doi.org/10.3390/ijms22147720
Chicago/Turabian StyleSalama, Abou Bakr M., Ahmad Gebreil, Tamer M. A. Mohamed, and Riham R. E. Abouleisa. 2021. "Induced Cardiomyocyte Proliferation: A Promising Approach to Cure Heart Failure" International Journal of Molecular Sciences 22, no. 14: 7720. https://doi.org/10.3390/ijms22147720