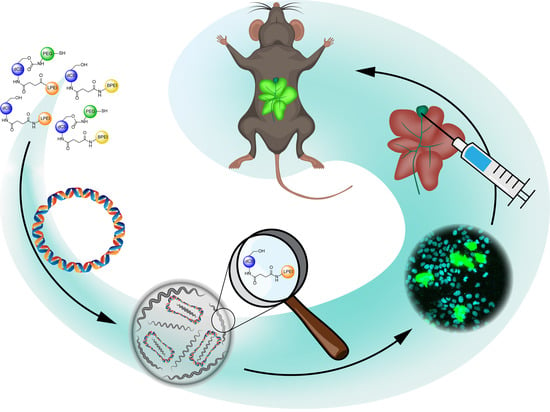
Development of Covalent Chitosan-Polyethylenimine Derivatives as Gene Delivery Vehicle: Synthesis, Characterization, and Evaluation
Abstract
:1. Introduction
2. Results
2.1. Synthesis and Physicochemical Properties of Covalent dCS-PEI Derivatives
2.2. NMR Analyses of Covalent dCS-PEI Derivatives
2.3. In Vitro Performance of DNA Complexed Covalent dCS-PEI Derivatives
2.4. Selection of a Lead Compound
2.5. In Vitro Gene Expression and Cytotoxicity Evaluation of the Lead Candidate
2.6. In Vivo Assessment of the Lead Candidate
3. Discussion
4. Materials and Methods
4.1. Materials and Cell Cultures
4.2. Polymers Analytics
4.3. Synthesis of Polymers
4.4. Polymer Complexation with DNA
4.5. Colloidal Stability
4.6. Transfection
4.7. Confocal Microscopy
4.8. Cytotoxicity
4.9. Retrograde Intrabiliary Infusion in Mice
4.10. In Vivo Imaging of Gene Expression and Assessment of Liver Specific Toxicity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 7-AAD | 7-Aminoactinomycin D |
| ALP | Alkaline phosphatase |
| ALT | Alkaline transaminase |
| BPEI | Branched polyethylenimine |
| CDI | 1,1′-carbonyldiimidazole |
| c/p | weight ratio of polymer [c] to nucleic acid [p] |
| CS | Chitosan |
| dCS | Depolymerized chitosan |
| DD | Deacetylation degree |
| DIPEA | N,N-diisopropylethylamine |
| DLS | Dynamic light scattering |
| DMSO | Dimethyl sulfoxide |
| DMTMM | 4-(4,6-dimethoxy-1,3,5-triazin-2-yl)-4-methyl-morpholinium chloride |
| DNA | Deoxyribonucleic acid |
| DOSY | Diffusion order spectroscopy |
| DSC | N-N’ disuccinimidyl carbonate |
| EDC | 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide |
| ELS | Electrophoretic light scattering |
| FACS | Fluorescence-activated cell sorting |
| FTIR | Fourier-transform infrared spectroscopy |
| GD | Grafting degree |
| GFP | Green fluorescent protein |
| GPC | Gel permeation chromatography |
| IVIS | In vivo Imaging system |
| LPEI | Linear polyethylenimine |
| Luc | Luciferase |
| Mn | Number average molecular weight |
| Mp | Peak molecular weight |
| MTS | 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium |
| MW | Weight average molecular weight |
| NHS | N-Hydroxysuccinimide |
| NMR | Nuclear magnetic resonance |
| NSuc | Suc linked to amine groups |
| OPEG | PEG linked to hydroxyl groups |
| PBS | Phosphate buffered saline |
| PDI | Polydispersity index |
| PEG | Polyethylene glycol |
| PEI | Polyethylenimine |
| pH | Potential of hydrogen |
| PLL | Poly-L-Lysine |
| rt | Room temperature |
| Suc | Succinate |
| w% | Weight percentage compared to total molecular weight |
References
- Mendell, J.R.; Al-Zaidy, S.A.; Rodino-Klapac, L.R.; Goodspeed, K.; Gray, S.J.; Kay, C.N.; Boye, S.L.; Boye, S.E.; George, L.A.; Salabarria, S.; et al. Current clinical applications of in vivo gene therapy with AAVs. Mol. Ther. 2021, 29, 464–488. [Google Scholar] [CrossRef]
- Dunbar, C.E.; High, K.A.; Joung, J.K.; Kohn, D.B.; Ozawa, K.; Sadelain, M. Gene therapy comes of age. Science 2018, 359, eaan4672. [Google Scholar] [CrossRef] [Green Version]
- Anzalone, A.V.; Koblan, L.W.; Liu, D.R. Genome Editing with CRISPR–Cas Nucleases, Base Editors, Transposases and prime editors. Nat. Biotech. 2020, 38, 824–844. [Google Scholar] [CrossRef]
- Santos-Carballal, B.; Fernández Fernández, E.; Goycoolea, F. Chitosan in Non-Viral Gene Delivery: Role of structure, characterization methods, and insights in cancer and rare diseases therapies. Polymers 2018, 10, 444. [Google Scholar] [CrossRef] [Green Version]
- Hardee, C.L.; Arévalo-Soliz, L.M.; Hornstein, B.D.; Zechiedrich, L. Advances in non-viral DNA vectors for gene therapy. Genes 2017, 8, 65. [Google Scholar] [CrossRef]
- Aravalli, R.N.; Belcher, J.D.; Steer, C.J. Liver-Targeted Gene therapy: Approaches and challenges. Liver Transplant. 2015, 21, 718–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibraheem, D.; Elaissari, A.; Fessi, H. Gene therapy and DNA delivery systems. Int. J. Pharm. 2014, 459, 70–83. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Zeng, X.; Liu, M.; Deng, Y.; He, N. Current Progress in Gene Delivery Technology based on chemical methods and nano-carriers. Theranostics 2014, 4, 240–255. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Xu, Y.; Wang, B.; Qiao, W.; Liu, D.; Li, Z. Cationic Compounds Used in lipoplexes and polyplexes for gene delivery. J. Control. Release 2004, 100, 165–180. [Google Scholar] [CrossRef] [PubMed]
- Kircheis, R.; Wightman, L.; Wagner, E. Design and Gene Delivery Activity of modified polyethylenimines. Adv. Drug Deliv. Rev. 2001, 53, 341–358. [Google Scholar] [CrossRef]
- Lungu, C.; Diudea, M.; Putz, M.; Grudziński, I. Linear and branched PEIs (Polyethylenimines) and their property space. Int. J. Mol. Sci. 2016, 17, 555. [Google Scholar] [CrossRef]
- Virgen-Ortíz, J.J.; Dos Santos, J.C.S.; Berenguer-Murcia, Á.; Barbosa, O.; Rodrigues, R.C.; Fernandez-Lafuente, R. Polyethylenimine: A very useful ionic polymer in the design of immobilized enzyme biocatalysts. J. Mater. Chem. B. 2017, 5, 7461–7490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, A.; Lächelt, U.; Bartek, J.; Wagner, E.; Moghimi, S.M. Polyplex Evolution: Understanding biology, optimizing performance. Mol Ther. 2017, 25, 1476–1490. [Google Scholar] [CrossRef] [Green Version]
- Boussif, O.; Lezoualc’ht, F.; Zanta, M.A.; Mergnyt, D.; Schermant, D.; Demeneixt, B.; Behr, J.-P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine. Proc. Natl. Acad. Sci. USA 1995, 92, 7297–7301. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; He, Z.; Hao, Y.; Gong, L.; Pang, M.; Howard, G.P.; Ahn, H.H.; Brummet, M.; Chen, K.; Liu, H.W.; et al. Kinetic control in assembly of plasmid DNA/polycation complex nanoparticles. ACS Nano. 2019, 13, 10161–10178. [Google Scholar] [CrossRef]
- Yen, J.; Ying, H.; Wang, H.; Yin, L.; Uckun, F.; Cheng, J. CD44 Mediated nonviral gene delivery into human embryonic stem cells via hyaluronic-acid-coated nanoparticles. ACS Biomater. Sci. Eng. 2016, 2, 326–335. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Cui, W.; Prasad, P.; Touve, M.A.; Gianneschi, N.C.; Mager, J.; Thayumanavan, S. Bait-and-switch supramolecular strategy to generate noncationic RNA-polymer complexes for RNA delivery. Biomacromolecules 2019, 20, 435–442. [Google Scholar] [CrossRef]
- Singh, B.; Maharjan, S.; Park, T.E.; Jiang, T.; Kang, S.K.; Choi, Y.J.; Cho, C.S. Tuning the buffering capacity of polyethylenimine with glycerol molecules for efficient gene delivery: Staying in or out of the endosomes. Macromol. Biosci. 2015, 15, 622–635. [Google Scholar] [CrossRef]
- Smith, S.A.; Selby, L.I.; Johnston, A.P.R.; Such, G.K. The Endosomal Escape of Nanoparticles: Toward more efficient cellular delivery. Bioconjug. Chem. 2019, 30, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Pei, D.; Buyanova, M. Overcoming Endosomal Entrapment in Drug Delivery. Bioconjug. Chem. 2019, 30, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.H.; Song, S.H.; Lim, D.W.; Lee, H.; Park, T.G. DNA transfection using linear poly(ethylenimine) prepared by controlled acid hydrolysis of poly(2-Ethyl-2-oxazoline). J. Control. Release 2001, 73, 391–399. [Google Scholar] [CrossRef]
- Neu, M.; Fischer, D.; Kissel, T. Recent Advances in Rational Gene Transfer Vector Design Based on Poly(Ethylene Imine) and Its Derivatives. J. Gene Med. 2005, 7, 992–1009. [Google Scholar] [CrossRef]
- Chen, J.; Wang, K.; Wu, J.; Tian, H.; Chen, X. Polycations for gene delivery: Dilemmas and solutions. Bioconjug. Chem. 2019, 30, 338–349. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, K.; Nishikawa, M.; Kawakami, S.; Nakano, T.; Hattori, Y.; Fumoto, S.; Yamashita, F.; Hashida, M. Molecular weight-dependent gene transfection activity of unmodified and galactosylated polyethyleneimine on hepatoma cells and mouse liver. Mol. Ther. 2003, 7, 254–261. [Google Scholar] [CrossRef]
- Wu, D.; Zhang, Y.; Xu, X.; Guo, T.; Xie, D.; Zhu, R.; Chen, S.; Ramakrishna, S.; He, L. RGD/TAT-functionalized chitosan-graft-PEI-PEG gene nanovector for sustained delivery of NT-3 for potential application in neural regeneration. Acta Biomater. 2018, 72, 266–277. [Google Scholar] [CrossRef]
- Riley, M.; Vermerris, W. Recent Advances in Nanomaterials for Gene Delivery—A Review. Nanomaterials 2017, 7, 94. [Google Scholar] [CrossRef] [Green Version]
- Mastorakos, P.; Zhang, C.; Berry, S.; Oh, Y.; Lee, S.; Eberhart, C.G.; Woodworth, G.F.; Suk, J.S.; Hanes, J. Highly PEGylated DNA nanoparticles provide uniform and widespread gene transfer in the brain. Adv. Healthc. Mater. 2015, 4, 1023–1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kunath, K.; Von Harpe, A.; Fischer, D.; Petersen, H.; Bickel, U.; Voigt, K.; Kissel, T. Low-molecular-weight polyethylenimine as a non-viral vector for DNA delivery: Comparison of physicochemical properties, transfection efficiency and in vivo distribution with high-molecular-weight polyethylenimine. J. Control. Release 2003, 89, 113–125. [Google Scholar] [CrossRef]
- Hill, A.B.; Chen, M.; Chen, C.K.; Pfeifer, B.A.; Jones, C.H. Overcoming gene-delivery hurdles: Physiological considerations for nonviral vectors. Trends Biotechnol. 2016, 34, 91–105. [Google Scholar] [CrossRef] [Green Version]
- Shahryari, A.; Jazi, M.S.; Mohammadi, S.; Nikoo, H.R.; Nazari, Z.; Hosseini, E.S.; Burtscher, I.; Mowla, S.J.; Lickert, H. Development and clinical translation of approved gene therapy products for genetic disorders. Front. Genet. 2019, 10, 868. [Google Scholar] [CrossRef] [Green Version]
- Mittal, H.; Ray, S.S.; Kaith, B.S.; Bhatia, J.K.; Sukriti; Sharma, J.; Alhassan, S.M. Recent progress in the structural modification of chitosan for applications in diversified biomedical fields. Eur. Polym. J. 2018, 109, 402–434. [Google Scholar] [CrossRef]
- Yadav, M.; Goswami, P.; Paritosh, K.; Kumar, M.; Pareek, N.; Vivekanand, V. Seafood waste: A source for preparation of commercially employable chitin/chitosan materials. Bioresour. Bioprocess. 2019, 6. [Google Scholar] [CrossRef]
- Radwan-Pragłowska, J.; Janus, Ł.; Piątkowski, M.; Bogdał, D.; Matysek, D. 3D Hierarchical, Nanostructured Chitosan/PLA/HA Scaffolds Doped with TiO2/Au/Pt NPs with Tunable Properties for Guided Bone Tissue Engineering. Polymers 2020, 12, 792. [Google Scholar] [CrossRef] [Green Version]
- Mora-Boza, A.; López-Ruiz, E.; López-Donaire, M.L.; Jiménez, G.; Aguilar, M.R.; Marchal, J.A.; Pedraz, J.L.; Vázquez-Lasa, B.; Román, J.S.; Gálvez-Martín, P. Evaluation of glycerylphytate crosslinked semi- and interpenetrated polymer membranes of hyaluronic acid and chitosan for tissue engineering. Polymers 2020, 12, 2661. [Google Scholar] [CrossRef]
- Yar, M.; Shahzad, S.; Shahzadi, L.; Shahzad, S.A.; Mahmood, N.; Chaudhry, A.A.; Rehman, I.U.; MacNeil, S. Heparin binding chitosan derivatives for production of pro-angiogenic hydrogels for promoting tissue healing. Mater. Sci. Eng. C 2017, 74, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cen, C.; Chen, J.; Fu, L. MgO/carboxymethyl chitosan nanocomposite improves thermal stability, waterproof and antibacterial performance for food packaging. Carbohydr. Polym. 2020, 236, 116078. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhu, Y.; Bai, Z.; Luque, R.; Xuan, J. Functionalized chitosan biosorbents with ultra-high performance, mechanical strength and tunable selectivity for heavy metals in wastewater treatment. Chem. Eng. J. 2017, 325, 350–359. [Google Scholar] [CrossRef]
- Yan, C.-Y.; Gu, J.-W.; Hou, D.-P.; Jing, H.-Y.; Wang, J.; Guo, Y.-Z.; Katsumi, H.; Sakane, T.; Yamamoto, A. synthesis of Tat tagged and folate modified N-succinyl-chitosan self-assembly nanoparticles as a novel gene vector. Int. J. Biol. Macromol. 2015, 72, 751–756. [Google Scholar] [CrossRef]
- Kong, F.; Tang, C.; Yin, C. Benzylguanidine and galactose double-conjugated chitosan nanoparticles with reduction responsiveness for targeted delivery of doxorubicin to CXCR 4 positive tumors. Bioconjug. Chem. 2020, 31, 2446–2455. [Google Scholar] [CrossRef] [PubMed]
- Kean, T.; Thanou, M. Biodegradation, biodistribution and toxicity of chitosan. Adv. Drug Deliv. Rev. 2010, 62, 3–11. [Google Scholar] [CrossRef]
- Thomas, T.; Tajmir-Riahi, H.; Pillai, C. Biodegradable polymers for gene delivery. Molecules 2019, 24, 3744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, Y.; Tan, Y.F.; Wong, Y.S.; Liew, M.W.J.; Venkatraman, S. Recent advances in chitosan-based carriers for gene delivery. Mar. Drugs 2019, 17, 381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavertu, M.; Méthot, S.; Tran-Khanh, N.; Buschmann, M.D. High efficiency gene transfer using chitosan/DNA nanoparticles with specific combinations of molecular weight and degree of deacetylation. Biomaterials 2006, 27, 4815–4824. [Google Scholar] [CrossRef] [PubMed]
- Kritchenkov, A.S.; Andranovitš, S.; Skorik, Y.A. Chitosan and its derivatives: Vectors in gene therapy. Russ. Chem. Rev. 2017, 86, 231. [Google Scholar] [CrossRef]
- Chuan, D.; Jin, T.; Fan, R.; Zhou, L.; Guo, G. Chitosan for gene delivery: Methods for improvement and applications. Adv. Colloid Interface Sci. 2019, 268, 25–38. [Google Scholar] [CrossRef]
- Zhang, X.; Duan, Y.; Wang, D.; Bian, F. Preparation of arginine modified PEI-conjugated chitosan copolymer for DNA delivery. Carbohydr. Polym. 2015, 122, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Zhu, M.; Liu, K.; Fan, H.; Zhao, W.; Mao, Y.; Zhang, Y. A biodegradable polyethylenimine-based vector modified by trifunctional peptide R18 for enhancing gene transfection efficiency in vivo. PLoS ONE 2016, 11, e0166673. [Google Scholar] [CrossRef]
- Journot, C.M.A.; Nicolle, L.; Lavanchy, Y.; Gerber-Lemaire, S. Selection of water-soluble chitosan by microwave-assisted degradation and pH-controlled precipitation. Polymers 2020, 12, 1274. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.Q.; Zhao, Q.Q.; Lv, T.F.; Shuai, W.P.; Zhou, J.; Tang, G.P.; Liang, W.Q.; Tabata, Y.; Hu, Y.L. Gene-carried chitosan-linked-PEI induced high gene transfection efficiency with low toxicity and significant tumor-suppressive activity. Int. J. Pharm. 2010, 387, 286–294. [Google Scholar] [CrossRef]
- Lu, H.; Dai, Y.; Lv, L.; Zhao, H. Chitosan-graft-polyethylenimine/DNA nanoparticles as novel non-viral gene delivery vectors targeting osteoarthritis. PLoS ONE 2014, 9, e84703. [Google Scholar] [CrossRef]
- Aiedeh, K.; Taha, M.O. Synthesis of chitosan succinate and chitosan phthalate and their evaluation as suggested matrices in orally administered, colon-specific drug delivery systems. Arch. Pharm. 1999, 332, 103–107. [Google Scholar] [CrossRef]
- Van Der Aa, M.A.E.M.; Huth, U.S.; Häfele, S.Y.; Schubert, R.; Oosting, R.S.; Mastrobattista, E.; Hennink, W.E.; Peschka-Süss, R.; Koning, G.A.; Crommelin, D.J.A. Cellular uptake of cationic polymer-DNA complexes via caveolae plays a pivotal role in gene transfection in COS-7 cells. Pharm. Res. 2007, 24, 1590–1598. [Google Scholar] [CrossRef] [Green Version]
- Zhu, M.; Nie, G.; Meng, H.; Xia, T.; Nel, A.; Zhao, Y. Physicochemical properties determine nanomaterial cellular uptake, transport, and fate. Acc. Chem. Res. 2013, 46, 622–631. [Google Scholar] [CrossRef] [Green Version]
- Hauck, T.S.; Ghazani, A.A.; Chan, W.C.W. Assessing the effect of surface chemistry on gold nanorod uptake, toxicity, and gene expression in mammalian cells. Small 2008, 4, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Danaei, M.; Dehghankhold, M.; Ataei, S.; Hasanzadeh Davarani, F.; Javanmard, R.; Dokhani, A.; Khorasani, S.; Mozafari, M.R. Impact of particle size and polydispersity index on the clinical applications of lipidic nanocarrier systems. Pharmaceutics 2018, 10, 57. [Google Scholar] [CrossRef] [Green Version]
- Bonnet, M.E.; Erbacher, P.; Bolcato-Bellemin, A.L. Systemic delivery of DNA or siRNA mediated by linear polyethylenimine (L-PEI) does not induce an inflammatory response. Pharm. Res. 2008, 25, 2972–2982. [Google Scholar] [CrossRef]
- Sadikot, R.T.; Jansen, E.D.; Blackwell, T.R.; Zoia, O.; Yull, F.; Christman, J.W.; Blackwell, T.S. High-dose dexamethasone accentuates nuclear factor-ΚB activation in endotoxin-treated mice. Am. J. Respir. Crit. Care Med. 2001, 164, 873–878. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; Sun, Y.X.; Li, Y.Q.; Zhang, X.Z.; Zhuo, R.X. N-succinyl-chitosan grafted with low molecular weight polyethylenimine as a serum-resistant gene vector. Mol. Biosyst. 2009, 5, 629–637. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.L.; Kim, Y.K.; Arote, R.; Nah, J.W.; Cho, M.H.; Choi, Y.J.; Akaike, T.; Cho, C.S. Chitosan-graft-polyethylenimine as a gene carrier. J. Control. Release 2007, 117, 273–280. [Google Scholar] [CrossRef]
- Lu, B.; Xu, X.D.; Zhang, X.Z.; Cheng, S.X.; Zhuo, R.X. Low molecular weight polyethylenimine grafted n-maleated chitosan for gene delivery: Properties and in vitro transfection studies. Biomacromolecules 2008, 9, 2594–2600. [Google Scholar] [CrossRef]
- Jiang, H.L.; Kim, Y.K.; Arote, R.; Jere, D.; Quan, J.S.; Yu, J.H.; Choi, Y.J.; Nah, J.W.; Cho, M.H.; Cho, C.S. Mannosylated chitosan-graft-polyethylenimine as a gene carrier for raw 264.7 cell targeting. Int. J. Pharm. 2009, 375, 133–139. [Google Scholar] [CrossRef]
- Chen, J.; Gao, X.; Hu, K.; Pang, Z.; Cai, J.; Li, J.; Wu, H.; Jiang, X. Galactose-poly(ethylene glycol)-polyethylenimine for improved lung gene transfer. Biochem. Biophys. Res. Commun. 2008, 375, 378–383. [Google Scholar] [CrossRef]
- Zhang, Z.; Yang, C.; Duan, Y.; Wang, Y.; Liu, J.; Wang, L.; Kong, D. Poly(ethylene glycol) analogs grafted with low molecular weight poly (ethylene imine) as non-viral gene vectors. Acta Biomater. 2010, 6, 2650–2657. [Google Scholar] [CrossRef] [PubMed]
- Chai, Z.; Zhang, X.; Dobbins, A.L.; Rigsbee, K.M.; Wang, B.; Samulski, R.J.; Li, C. Optimization of dexamethasone administration for maintaining global transduction efficacy of adeno-associated virus serotype 9. Hum. Gene Ther. 2019, 30, 829–840. [Google Scholar] [CrossRef] [PubMed]
- Seregin, S.S.; Appledorn, D.M.; McBride, A.J.; Schuldt, N.J.; Aldhamen, Y.A.; Voss, T.; Wei, J.; Bujold, M.; Nance, W.; Godbehere, S.; et al. Transient pretreatment with glucocorticoid ablates innate toxicity of systemically delivered adenoviral vectors without reducing efficacy. Mol. Ther. 2009, 17, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Suhr, O.B.; Coelho, T.; Buades, J.; Pouget, J.; Conceicao, I.; Berk, J.; Schmidt, H.; Waddington-Cruz, M.; Campistol, J.M.; Bettencourt, B.R.; et al. Efficacy and safety of patisiran for familial amyloidotic polyneuropathy: A phase II multi-dose study. Orphanet J. Rare Dis. 2015, 10, 109. [Google Scholar] [CrossRef] [Green Version]
- Nair, N.; Rincon, M.Y.; Evens, H.; Sarcar, S.; Dastidar, S.; Samara-Kuko, E.; Ghandeharian, O.; Viecelli, H.M.; Thöny, B.; De Bleser, P.; et al. Computationally designed liver-specific transcriptional modules and hyperactive factor ix improve hepatic gene therapy. Blood 2014, 123, 3195–3199. [Google Scholar] [CrossRef] [Green Version]
- Viecelli, H.M.; Harbottle, R.P.; Wong, S.P.; Schlegel, A.; Chuah, M.K.; Vandendriessche, T.; Harding, C.O.; Thöny, B. Treatment of phenylketonuria using minicircle-based naked-DNA gene transfer to murine liver. Hepatology 2014, 60, 1035–1043. [Google Scholar] [CrossRef] [Green Version]
- Grisch-Chan, H.M.; Schlegel, A.; Scherer, T.; Allegri, G.; Heidelberger, R.; Tsikrika, P.; Schmeer, M.; Schleef, M.; Harding, C.O.; Häberle, J.; et al. Low-dose gene therapy for murine PKU using episomal naked DNA vectors expressing PAH from its endogenous liver promoter. Mol. Ther. Nucleic Acids 2017, 7, 339–349. [Google Scholar] [CrossRef] [Green Version]
- Dai, H.; Jiang, X.; Tan, G.C.Y.; Chen, Y.; Torbenson, M.; Leong, K.W.; Mao, H.Q. Chitosan-DNA nanoparticles delivered by intrabiliary infusion enhance liver-targeted gene delivery. Int. J. Nanomed. 2006, 1, 507–522. [Google Scholar] [CrossRef] [Green Version]
- Jiang, X.; Dai, H.; Ke, C.Y.; Mo, X.; Torbenson, M.S.; Li, Z.; Mao, H.Q. PEG-b-PPA/DNA micelles improve transgene expression in rat liver through intrabiliary infusion. J. Control. Release 2007, 122, 297–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allan, C.; Burel, J.M.; Moore, J.; Blackburn, C.; Linkert, M.; Loynton, S.; MacDonald, D.; Moore, W.J.; Neves, C.; Patterson, A.; et al. OMERO: Flexible, model-driven data management for experimental biology. Nat. Methods. 2012, 9, 245–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berntsen, N.L.; Fosby, B.; Valestrand, L.; Tan, C.; Reims, H.M.; Schrumpf, E.; Karlsen, T.H.; Line, P.D.; Melum, E. Establishment of a surgical bile duct injection technique giving direct access to the bile ducts for studies of the murine biliary tree. Am. J. Physiol. Gastrointest. Liver Physiol. 2018, 314, G349–G359. [Google Scholar] [CrossRef] [PubMed]





| Polymer | Total Molecular Weight (Da) | Depolymerized Chitosan (dCS) | Succinate (Suc) | Polyethylenimine (PEI) | Polyethylene Glycol (PEG) | Solubility 1 (mg/mL) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mw (Da) | DD (%) | GD (%) | w% | GD (%) | w% | GD (%) | w% | |||
Starting polymer
| ||||||||||
| 8300 | 8300 | 79 | - | - | - | - | - | - | 3 | |
| 2500 | - | - | - | - | - | - | - | - | 5 | |
| 1800 | - | - | - | - | - | - | - | - | >10 | |
BPEI derivatives
| ||||||||||
| 19,100 | 7800 | 80 | 47 | 12 | 11 | 48 | - | - | >10 | |
| 16,900 | 6200 | 80 | 54 | 12 | 13 | 51 | - | - | 7.5 | |
| 75,300 | 9000 | 80 | 44 | 3 | 67 | 85 | - | - | >10 | |
| 43,500 | 7800 | 80 | 47 | 5 | 11 | 21 | 26 | 56 | >10 | |
LPEI derivatives 2
| ||||||||||
| 16,800 | 5700 | 80 | 51 | 10 | 11 | 56 | - | - | >10 | |
| 23,800 | 7800 | 80 | 47 | 10 | 11 | 56 | - | - | >10 | |
| 28,600 | 8300 | 79 | 63 | 11 | 14 | 60 | - | - | >10 | |
| 48,300 | 7800 | 85 | 44 | 4 | 22 | 53 | 14 | 27 | >10 | |
| Polymers | Polyplex Stability (c/p Ratio) | Optimized Colloidal Stability (c/p Ratio) | ζ-Potential (mV) | Hydrodynamic Diameter D (nm) | PDI | Luc x GFP Normalized to dCS c/p 32 (Fold) | MTS (% Viable Cells) |
|---|---|---|---|---|---|---|---|
Starting polymer
| |||||||
| 32 | 32 | 22 4 | 68 5 | 0.23 0.03 | 1 1 | 111 5 | |
| 32 | 32 | 28 7 | 48 1 | 0.17 0.01 | 2103 61 | 25 1 | |
| 16 | 16 | 21 1 | 91 1 | 0.09 0.02 | 3415 119 | 43 4 | |
BPEI derivatives
| |||||||
| 1 | 2 | 19 3 | 68 2 | 0.22 0.01 | 415 24 | 81 1 | |
| 0.5 | 4 | 16 4 | 62 1 | 0.22 0.01 | 1174 6 | 81 7 | |
| 0.5 | 1 | 24 2 | 79 ± 1 | 0.21 0.01 | 3488 160 | 63 2 | |
| 1 | 2 | 13 1 | 121 1 | 0.21 0.01 | 0 0 | 91 2 | |
LPEI derivatives
| |||||||
| 1 | 1 | 17 5 | 74 2 | 0.25 0.02 | 3076 73 | 97 5 | |
| 2 | 2 | 23 1 | 87 1 | 0.16 0.01 | 71,567 ± 1592 | 101 9 | |
| 2 | 2 | 18 1 | 85 1 | 0.17 0.01 | 0 0 | 91 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nicolle, L.; Casper, J.; Willimann, M.; Journot, C.M.A.; Detampel, P.; Einfalt, T.; Grisch-Chan, H.M.; Thöny, B.; Gerber-Lemaire, S.; Huwyler, J. Development of Covalent Chitosan-Polyethylenimine Derivatives as Gene Delivery Vehicle: Synthesis, Characterization, and Evaluation. Int. J. Mol. Sci. 2021, 22, 3828. https://doi.org/10.3390/ijms22083828
Nicolle L, Casper J, Willimann M, Journot CMA, Detampel P, Einfalt T, Grisch-Chan HM, Thöny B, Gerber-Lemaire S, Huwyler J. Development of Covalent Chitosan-Polyethylenimine Derivatives as Gene Delivery Vehicle: Synthesis, Characterization, and Evaluation. International Journal of Molecular Sciences. 2021; 22(8):3828. https://doi.org/10.3390/ijms22083828
Chicago/Turabian StyleNicolle, Laura, Jens Casper, Melanie Willimann, Céline M. A. Journot, Pascal Detampel, Tomaž Einfalt, Hiu Man Grisch-Chan, Beat Thöny, Sandrine Gerber-Lemaire, and Jörg Huwyler. 2021. "Development of Covalent Chitosan-Polyethylenimine Derivatives as Gene Delivery Vehicle: Synthesis, Characterization, and Evaluation" International Journal of Molecular Sciences 22, no. 8: 3828. https://doi.org/10.3390/ijms22083828