The Effect of Virulence and Resistance Mechanisms on the Interactions between Parasitic Plants and Their Hosts
Abstract
:1. Introduction
2. Virulence Evolution in the Family Orobanchaceae
2.1. Definition of Race in Parasitic Plants
2.2. History of Race in Parasitic Plants
3. The Mechanisms of Virulence-Specific Resistance in Host Plants
3.1. Phenotypic Aspects
3.2. Histological Aspects
3.3. Chemical Aspects
3.4. Genetic and Genomic Aspects
3.4.1. R Genes against Orobanche cumana
3.4.2. R Genes against Striga gesnerioides
3.4.3. Virulence Genes in Parasitic Plants
3.4.4. Genome and Transcriptome of Parasitic Plants
4. Models of Interaction and Co-Evolution between Parasitic Plants and Their Hosts
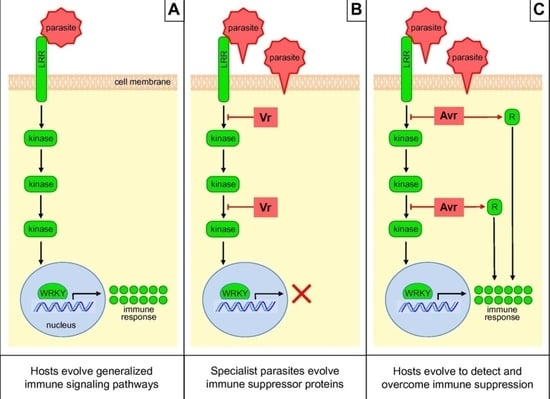
4.1. Model of Defense Activated by Host NLR Proteins Triggered by the Parasitic Plant
4.2. Model of Antagonistic Host–Parasite Co-Evolution
4.3. Drivers of Pathogen Effector Evolution
5. The Effect on Parasitic Plants
The Effect on Host Plants
6. Conclusions
Funding
Conflicts of Interest
Abbreviations
| HIGS | Host-induced gene silencing |
| HGT | Horizontal gene transfer |
| LRR | Leucine-rich repeat |
| NLR | Nucleotide-binding and leucine-rich repeat domain |
| PR | Pathogen-related protein gene |
| QTL | Quantitative trait loci |
| R | Resistance gene |
| SiRNA | Small interfering RNA |
| SL | Strigolactone |
| HGT | Horizontal gene transfer |
| GR24 | Synthetic analog of strigolactones |
| WRKY | Zinc-finger transcription factor-related domain containing the WRKY sequence |
| TF | Transcription factor |
| Avr | Avirulence |
| Vr | Virulence |
| RNAi | RNA interference |
| Crispr/Cas9 | Clustered regularly interspaced short palindromic repeats/CRISPR associated protein 9 |
References
- Yoshida, S.; Cui, S.; Ichihashi, Y.; Shirasu, K. The haustorium, a specialized invasive organ in parasitic plants. Annu. Rev. Plant. Biol. 2016, 67, 643–667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heide-Jørgensen, H.S. Introduction: The parasitic syndrome in higher plants. In Parasitic Orobanchaceae; Springer: Berlin/Heidelberg, Germany, 2013; pp. 1–18. [Google Scholar]
- Westwood, J.H.; Yoder, J.I.; Timko, M.P.; dePamphilis, C.W. The evolution of parasitism in plants. Trends Plant. Sci. 2010, 15, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Skippington, E.; Barkman, T.J.; Rice, D.W.; Palmer, J.D. Comparative mitogenomics indicates respiratory competence in parasitic Viscum despite loss of complex I and extreme sequence divergence, and reveals horizontal gene transfer and remarkable variation in genome size. BMC Plant. Biol. 2017, 17, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parker, C. Observations on the current status of Orobanche and Striga problems worldwide. Pest. Manag. Sci. 2009, 65, 453–459. [Google Scholar] [CrossRef]
- Fernández-Aparicio, M.; Reboud, X.; Gibot-Leclerc, S. Broomrape weeds. Underground mechanisms of parasitism and associated strategies for their control: A review. Front. Plant. Sci. 2016, 7, 135. [Google Scholar] [CrossRef] [PubMed]
- Bouwmeester, H.J.; Matusova, R.; Zhongkui, S.; Beale, M.H. Secondary metabolite signalling in host-parasitic plant interactions. Curr. Opin. Plant. Biol. 2003, 6, 358–364. [Google Scholar] [CrossRef]
- Zhou, W.J.; Yoneyama, K.; Takeuchi, Y.; Iso, S.; Rungmekarat, S.; Chae, S.H.; Joel, D.M. In vitro infection of host roots by differentiated calli of the parasitic plant Orobanche. J. Exp. Bot. 2004, 55, 899–907. [Google Scholar] [CrossRef] [Green Version]
- Matusova, R.; Rani, K.; Verstappen, F.W.A.; Franssen, M.C.R.; Beale, M.H.; Bouwmeester, H.J. The strigolactone germination stimulants of the plant-parasitic Striga and Orobanche spp. are derived from the carotenoid pathway. Plant. Physiol. 2005, 139, 920–934. [Google Scholar] [CrossRef] [Green Version]
- Screpanti, C.; Yoneyama, K.; Bouwmeester, H.J. Strigolactones and parasitic weed management 50 years after the discovery of the first natural strigolactone strigol: Status and outlook. Pest. Manag. Sci. 2016, 72, 2013–2015. [Google Scholar] [CrossRef]
- Ejeta, G. Breeding for Striga resistance in Sorghum: Exploitation of an intricate host-parasite biology. Crop. Sci. 2007, 47, 216–227. [Google Scholar] [CrossRef]
- Fernández-Aparicio, M.; Kisugi, T.; Xie, X.; Rubiales, D.; Yoneyama, K. Low strigolactone root exudation: A novel nechanism of Broomrape (Orobanche and Phelipanche spp.) resistance available forfaba bean breeding. J. Agric. Food Chem. 2014, 62, 7063–7071. [Google Scholar] [CrossRef] [PubMed]
- Jamil, M.; Rodenburg, J.; Charnikhova, T.; Bouwmeester, H.J. Pre-attachment Striga hermonthica resistance of New Rice for Africa (NERICA) cultivars based on low strigolactone production. New Phytol. 2011, 192, 964–975. [Google Scholar] [CrossRef] [PubMed]
- Pavan, S.; Schiavulli, A.; Marcotrigiano, A.R.; Bardaro, N.; Bracuto, V.; Ricciardi, F.; Charnikhova, T.; Lotti, C.; Bouwmeester, H.; Ricciardi, L. Characterization of low-strigolactone germplasm in pea (Pisum sativum L.) resistant to crenate broomrape (Orobanche crenata Forsk.). Mol. Plant. Microbe. Interact. 2016, 29, 743–749. [Google Scholar] [CrossRef] [Green Version]
- Gevezova, M.; Dekalska, T.; Stoyanov, K.; Hristeva, T.; Kostov, K.; Batchvarova, R.; Denev, I. Recent advances in broomrapes research. J. Biosci. Biotechnol. 2012, 91–105. [Google Scholar]
- Samejima, H.; Sugimoto, Y. Recent research progress in combatting root parasitic weeds. Biotechnol. Biotechnol. Equip. 2018, 32, 221–240. [Google Scholar] [CrossRef] [Green Version]
- Islam, F.; Wang, J.; Farooq, M.A.; Khan, M.S.; Xu, L.; Zhu, J.; Zhou, W. Potential impact of the herbicide 2,4-dichlorophenoxyacetic acid on human and ecosystems. Environ. Intl. 2018, 111, 332–351. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, J.; DiTommaso, A.; Zhang, C.; Zheng, G.; Liang, W.; Zhou, W. Weed research status, challenges, and opportunities in China. Crop. Prot. 2020, 134, 104449. [Google Scholar] [CrossRef]
- Molinero-Ruiz, L.; Delavault, P.; Pérez-Vich, B.; Pacureanu-Joita, M.; Bulos, M.; Altieri, E.; Domínguez, J. History of the race structure of Orobanche cumana and the breeding of sunflower for resistance to this parasitic weed: A review. Spanish J. Agric. Res. 2015, 13, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Louarn, J.; Boniface, M.-C.; Pouilly, N.; Velasco, L.; Pérez-Vich, B.; Vincourt, P.; Muños, S. Sunflower resistance to broomrape (Orobanche cumana) is controlled by specific QTLs for different parasitism stages. Front. Plant. Sci. 2016, 7, 590. [Google Scholar] [CrossRef] [Green Version]
- Alcántara, E.; Morales-García, M.; Díaz-Sánchez, J. Effects of broomrape parasitism on sunflower plants: Growth, development, and mineral nutrition. J. Plant. Nutr. 2006, 29, 1199–1206. [Google Scholar] [CrossRef]
- Duca, M. Historical aspects of sunflower researches in the Republic of Moldova. Helia 2015, 38, 79–92. [Google Scholar] [CrossRef]
- Yoder, J.I.; Scholes, J.D. Host plant resistance to parasitic weeds; recent progress and bottlenecks. Curr. Opin. Plant. Biol. 2010, 13, 478–484. [Google Scholar] [CrossRef]
- Botanga, C.J.; Timko, M.P. Phenetic relationships among different races of Striga gesnerioides (Willd.) Vatke from West Africa. Genome 2006, 49, 1351–1365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antonova, T.S.; Araslanova, N.M.; Strelnikov, E.A.; Ramazanova, S.A.; Guchetl, S.Z.; Chelyustnikova, T.A. Distribution of highly virulent races of sunflower broomrape (Orobanche cumana Wallr.) in the southern regions of the Russian Federation. Russ. Agric. Sci. 2013, 39, 46–50. [Google Scholar] [CrossRef]
- Delipavlov, D. Orobanchaceae. In Flora Reipublicae Bulgaricae; Kozuharov, S.I., Kuzmanov, B.A., Eds.; Marin Drinov Academic Publishing House: Sofia, Bulgaria, 1995; Volume 10, pp. 291–325. [Google Scholar]
- Antonova, T.S. The history of interconnected evolution of Orobanche cumana Wallr. and sunflower in the Russian Federation and Kazakhstan. Helia 2014, 37, 215–225. [Google Scholar] [CrossRef] [Green Version]
- Shi, B.; Lei, Z.; Xiang, L.; Zhao, J. Identification of sunflower as physiological species in 4 provinces of China. Chin. J. Oil Crops Sci. 2016, 38, 116–119. [Google Scholar] [CrossRef]
- Shi, B.X.; Chen, G.H.; Zhang, Z.J.; Hao, J.J.; Jing, L.; Zhou, H.Y.; Zhao, J. First report of race composition and distribution of sunflower broomrape, Orobanche cumana, in China. Plant. Dis. 2015, 99, 291. [Google Scholar] [CrossRef] [PubMed]
- Nabloussi, A.; Velasco, L.; Assissel, N. First report of sunflower broomrape, Orobanche cumana Wallr., in Morocco. Plant. Dis. 2018, 102, 457. [Google Scholar] [CrossRef]
- González-Cantón, E.; Velasco, A.; Velasco, L.; Pérez-Vich, B.; Martín-Sanz, A. First report of sunflower broomrape (Orobanche cumana) in Portugal. Plant. Dis. 2019, 8, 2143. [Google Scholar] [CrossRef]
- Pérez de Luque, A.; Jorrín, J.; Cubero, J.I.; Rubiales, D. Orobanche crenata resistance and avoidance in pea (Pisum spp.) operate at different developmental stages of the parasite. Weed Res. 2005, 45, 379–387. [Google Scholar] [CrossRef]
- Rubiales, D.; Rojas-Molina, M.M.; Sillero, J.C. Characterization of resistance mechanisms in faba bean (Vicia faba) against broomrape species (Orobanche and Phelipanche spp.). Front. Plant. Sci. 2016, 7, 1747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martín-Sanz, A.; Pérez-Vich, B.; Rueda, S.; Fernández-Martínez, J.M.; Velasco, L. Characterization of post-haustorial resistance to sunflower broomrape. Crop. Sci. 2020, 60, 1188–1198. [Google Scholar] [CrossRef]
- Rodenburg, J.; Cissoko, M.; Kayongo, N.; Dieng, I.; Bisikwa, J.; Irakiza, R.; Masoka, I.; Midega, C.A.O.; Scholes, J.D. Genetic variation and host-parasite specificity of Striga resistance and tolerance in rice: The need for predictive breeding. New Phytol. 2017, 214, 1267–1280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cochavi, A.; Rapaport, T.; Gendler, T.; Karnieli, A.; Eizenberg, H.; Rachmilevitch, S.; Ephrath, J.E. Recognition of Orobanche cumana below-ground parasitism through physiological and hyper spectral measurements in sunflower (Helianthus annuus L.). Front. Plant. Sci. 2017, 8, 909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pincovici, S.; Cochavi, A.; Karnieli, A.; Ephrath, J.; Rachmilevitch, S. Source-sink relations of sunflower plants as affected by a parasite modifies carbon allocations and leaf traits. Plant. Sci. 2018, 271, 100–107. [Google Scholar] [CrossRef]
- Serghini, K.; Pérez de Luque, A.; Castejón-Muñoz, M.; García-Torres, L.; Jorrín, J. V Sunflower (Helianthus annuus L.) response to broomrape (Orobanche cernua Loefl.) parasitism: Induced synthesis and excretion of 7-hydroxylated simple coumarins. J. Exp. Bot. 2001, 52, 2227–2234. [Google Scholar] [CrossRef]
- Pérez-DE-Luque, A.; Rubiales, D.; Cubero, J.I.; Press, M.C.; Scholes, J.; Yoneyama, K.; Takeuchi, Y.; Plakhine, D.; Joel, D.M. Interaction between Orobanche crenata and its host legumes: Unsuccessful haustorial penetration and necrosis of the developing parasite. Ann. Bot. 2005, 95, 935–942. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Xu, L.; Zhang, N.; Islam, F.; Song, W.; Hu, L.; Liu, D.; Xie, X.; Zhou, W. iTRAQ-based proteomics of sunflower cultivars differing in resistance to parasitic weed Orobanche cumana. Proteomics 2017, 17, 1700009. [Google Scholar] [CrossRef]
- Mutuku, J.M.; Cui, S.; Hori, C.; Takeda, Y.; Tobimatsu, Y.; Nakabayashi, R.; Mori, T.; Saito, K.; Demura, T.; Umezawa, T.; et al. The structural integrity of lignin is crucial for resistance against Striga hermonthica parasitism in rice. Plant. Physiol. 2019, 179, 1796–1809. [Google Scholar] [CrossRef] [Green Version]
- Huang, K.; Mellor, K.E.; Paul, S.N.; Lawson, M.J.; Mackey, A.J.; Timko, M.P. Global changes in gene expression during compatible and incompatible interactions of cowpea (Vigna unguiculata L.) with the root parasitic angiosperm Striga gesnerioides. BMC Genomics 2012, 13, 1. [Google Scholar] [CrossRef] [Green Version]
- Pérez-de-Luque, A.; Moreno, M.T.; Rubiales, D. Host plant resistance against broomrapes (Orobanche spp.): Defence reactions and mechanisms of resistance. Ann. Appl. Biol. 2008, 152, 131–141. [Google Scholar] [CrossRef]
- Yoshida, S.; Shirasu, K. Multiple layers of incompatibility to the parasitic witchweed, Striga hermonthica. New Phytol. 2009, 183, 180–189. [Google Scholar] [CrossRef] [PubMed]
- Gurney, A.L.; Slate, J.; Press, M.C.; Scholes, J.D. A novel form of resistance in rice to the angiosperm parasite Striga hermonthica. New Phytol. 2006, 169, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Duriez, P.; Vautrin, S.; Auriac, M.C.; Bazerque, J.; Boniface, M.C.; Callot, C.; Carrère, S.; Cauet, S.; Chabaud, M.; Gentou, F.; et al. A receptor-like kinase enhances sunflower resistance to Orobanche cumana. Nat. Plants 2019, 5, 1211–1215. [Google Scholar] [CrossRef]
- López-Ráez, J.A.; Matusova, R.; Cardoso, C.; Jamil, M.; Charnikhova, T.; Kohlen, W.; Ruyter-Spira, C.; Verstappen, F.; Bouwmeester, H. Strigolactones: Ecological significance and use as a target for parasitic plant control. Pest. Manag. Sci. 2009, 65, 471–477. [Google Scholar] [CrossRef]
- Rubiales, D.; Pérez-de-Luque, A.; Joel, D.M.; Alcántara, C.; Sillero, J.C. Characterization of resistance in chickpea to crenate broomrape (Orobanche crenata). Weed Sci. 2003, 51, 702–707. [Google Scholar] [CrossRef]
- Rubiales, D.; Alcántara, C.; Pérez-de-Luque, A.; Gil, J.; Sillero, J.C. Infection of chickpea (Cicer arietinum) by crenate broomrape (Orobanche crenata) as influenced by sowing date and weather conditions. Agronomie 2003, 23, 359–362. [Google Scholar] [CrossRef]
- Sillero, J.C.; Cubero, J.I.; Fernández-Aparicio, M.; Rubiales, D. Search for resistance to crenate broomrape (Orobanche crenata). Lathyrus Lathyrism Newsl. 2005, 4, 7–9. [Google Scholar]
- Sillero, J.C.; Moreno, M.T.; Rubiales, D. Sources of resistance to crenate broomrape in Vicia species. Plant. Dis. 2005, 89, 23–27. [Google Scholar] [CrossRef] [Green Version]
- Abbes, Z.; Kharrat, M.; Pouvreau, J.B.; Delavault, P.; Chaibi, W.; Simier, P. The dynamics of faba bean (Vicia faba L.) parasitism by Orobanche foetida. Phytopathol. Mediterr. 2010, 49, 239–248. [Google Scholar]
- Jorrín, J.; Pérez-de-Luque, A.; Serghini, K.; Cubero, J.I.; Moreno, M.T.; Rubiales, D.; Sillero, J.C. Resistance to Orobanche: The State of the Art; Junta de Andalucía: Sevilla, Spain, 1999; Volume 51, pp. 163–177.
- Labrousse, P. Several mechanisms are involved in resistance of Helianthus to Orobanche cumana Wallr. Ann. Bot. 2001, 88, 859–868. [Google Scholar] [CrossRef] [Green Version]
- Labrousse, P.; Arnaud, M.C.; Griveau, Y.; Fer, A.; Thalouran, P. Analysis of resistance criteria of sunflower recombined inbred lines against Orobanche cumana Wallr. Crop. Prot. 2004, 23, 407–413. [Google Scholar] [CrossRef]
- Pierce, S.; Mbwaga, A.M.; Press, M.C.; Scholes, J.D. Xenognosin production and tolerance to Striga asiatica infection of high-yielding maize cultivars. Weed Res. 2003, 43, 139–145. [Google Scholar] [CrossRef]
- Fernández-Aparicio, M.; Moral, A.; Kharrat, M.; Rubiales, D. Resistance against broomrapes (Orobanche and Phelipanche spp.) in faba bean (Vicia faba) based in low induction of broomrape seed germination. Euphytica 2012, 186, 897–905. [Google Scholar] [CrossRef]
- Al-Babili, S.; Bouwmeester, H.J. Strigolactones, a novel carotenoid-derived plant hormone. Annu. Rev. Plant. Biol. 2015, 66, 161–186. [Google Scholar] [CrossRef]
- Cook, C.E.; Whichard, L.P.; Turner, B.; Wall, M.E.; Egley, G.H. Germination of Witchweed (Striga lutea Lour.): Isolationand properties of a potent stimulant. Science 1966, 154, 1189–1190. [Google Scholar] [CrossRef]
- Akiyama, K.; Matsuzaki, K.I.; Hayashi, H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 2005, 435, 824–827. [Google Scholar] [CrossRef]
- Besserer, A.; Puech-Pagès, V.; Kiefer, P.; Gomez-Roldan, V.; Jauneau, A.; Roy, S.; Portais, J.C.; Roux, C.; Bécard, G.; Séjalon-Delmas, N. Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol. 2006, 4, 1239–1247. [Google Scholar] [CrossRef]
- Gomez-Roldan, V.; Fermas, S.; Brewer, P.B.; Puech-Pagès, V.; Dun, E.A.; Pillot, J.P.; Letisse, F.; Matusova, R.; Danoun, S.; Portais, J.C.; et al. Strigolactone inhibition of shoot branching. Nature 2008, 455, 189–194. [Google Scholar] [CrossRef]
- Umehara, M.; Hanada, A.; Yoshida, S.; Akiyama, K.; Arite, T.; Takeda-Kamiya, N.; Magome, H.; Kamiya, Y.; Shirasu, K.; Yoneyama, K.; et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature 2008, 455, 195–200. [Google Scholar] [CrossRef]
- Alder, A.; Jamil, M.; Marzorati, M.; Bruno, M.; Vermathen, M.; Bigler, P.; Ghisla, S.; Bouwmeester, H.; Beyer, P.; Al-Babili, S. The path from β-Carotene to carlactone, a strigolactone-like plant hormone. Science 2012, 335, 1348–1351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruno, M.; Al-Babili, S. On the substrate specificity of the rice strigolactone biosynthesis enzyme DWARF27. Planta 2016, 243, 1429–1440. [Google Scholar] [CrossRef] [PubMed]
- Bruno, M.; Hofmann, M.; Vermathen, M.; Alder, A.; Beyer, P.; Al-Babili, S. On the substrate- and stereospecificity of the plant carotenoid cleavage dioxygenase 7. FEBS Lett. 2014, 588, 1802–1807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruno, M.; Vermathen, M.; Alder, A.; Wüst, F.; Schaub, P.; van der Steen, R.; Beyer, P.; Ghisla, S.; Al-Babili, S. Insights into the formation of carlactone from in-depth analysis of the CCD8-catalyzed reactions. FEBS Lett. 2017, 591, 792–800. [Google Scholar] [CrossRef]
- Jia, K.-P.; Baz, L.; Al-Babili, S. From carotenoids to strigolactones. J. Exp. Bot. 2017, 69, 2189–2204. [Google Scholar] [CrossRef] [Green Version]
- Waters, M.T.; Gutjahr, C.; Bennett, T.; Nelson, D.C. Strigolactone Signaling and Evolution. Annu. Rev. Plant. Biol. 2017, 68, 291–322. [Google Scholar] [CrossRef]
- Brewer, P.B.; Yoneyama, K.; Filardo, F.; Meyers, E.; Scaffidi, A.; Frickey, T.; Akiyama, K.; Seto, Y.; Dun, E.A.; Cremer, J.E.; et al. LATERAL BRANCHING OXIDOREDUCTASE acts in the final stages of strigolactone biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 6301–6306. [Google Scholar] [CrossRef] [Green Version]
- Abe, S.; Sado, A.; Tanaka, K.; Kisugi, T.; Asami, K.; Ota, S.; Kim, H., II; Yoneyama, K.; Xie, X.; Ohnishi, T.; et al. Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. Proc. Natl. Acad. Sci. USA 2014, 111, 18084–18089. [Google Scholar] [CrossRef] [Green Version]
- Iseki, M.; Shida, K.; Kuwabara, K.; Wakabayashi, T.; Mizutani, M.; Takikawa, H.; Sugimoto, Y. Evidence for species-dependent biosynthetic pathways for converting carlactone to strigolactones in plants. J. Exp. Bot. 2018, 69, 2305–2318. [Google Scholar] [CrossRef]
- Zhang, Y.; van Dijik, A.D.; Scaffidi, A.; Flematti, G.R.; Hofmann, M.; Charnikhova, T.; Verstappen, F.; Hepworth, J.; van der Krol, S.; Leyser, O.; et al. Rice cytochrome P450 MAX1 homologs catalyze distinct steps in strigolactone biosynthesis. Nat. Chem. Biol. 2014, 12, 1028–1033. [Google Scholar] [CrossRef]
- Pouvreau, J.B.; Gaudin, Z.; Auger, B.; Lechat, M.M.; Gauthier, M.; Delavault, P.; Simier, P. A high-throughput seed germination assay for root parasitic plants. Plant. Methods 2013, 9, 32. [Google Scholar] [CrossRef] [Green Version]
- Matusova, R.; van Mourik, T.; Bouwmeester, H.J. Changes in the sensitivity of parasitic weed seeds to germination stimulants. Seed Sci. Res. 2004, 14, 335–344. [Google Scholar] [CrossRef]
- Ueno, K.; Furumoto, T.; Umeda, S.; Mizutani, M.; Takikawa, H.; Batchvarova, R.; Sugimoto, Y. Heliolactone, a non-sesquiterpene lactone germination stimulant for root parasitic weeds from sunflower. Phytochemistry 2014, 108, 122–128. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Yang, C.; Liu, H.; Cao, M.; Yan, G.; Si, P.; Zhou, W.; Xu, L. 5-Aminolevolinic acid enhances sunflower resistance to Orobanche cumana (broomrape). Ind. Crops Prod. 2019, 140, 111467. [Google Scholar] [CrossRef]
- Yang, C.; Hu, L.Y.; Ali, B.; Islam, F.; Bai, Q.J.; Yun, X.P.; Zhou, W.J. Seed treatment with salicylic acid invokes defense mechanism of Helianthus annuus against Orobanche cumana. Ans. App. Biol. 2016, 169, 408–422. [Google Scholar] [CrossRef]
- Song, W.J.; Zhou, W.J.; Jin, Z.L.; Zhang, D.; Yoneyama, K.; Takeuchi, Y.; Joel, D.M. Growth regulators restore germination of Orobanche seeds that are conditioned under water stress and suboptimal temperature. Aus. J. Agri. Res. 2006, 57, 1195–1201. [Google Scholar] [CrossRef]
- Song, W.J.; Zhou, W.J.; Jin, Z.L.; Cao, D.D.; Joel, D.M.; Takeuchi, Y.; Yoneyama, K. Germination response of Orobanche seeds subjected to conditioning temperature, water potential and growth regulator treatments. Weed Res. 2005, 45, 467–476. [Google Scholar] [CrossRef]
- Yoneyama, K. Recent progress in the chemistry and biochemistry of strigolactones. J. Pestic. Sci. 2020, 45, 45–53. [Google Scholar] [CrossRef] [Green Version]
- Yoneyama, K.; Xie, X.; Yoneyama, K.; Nomura, T.; Takahashi, I.; Asami, T.; Mori, N.; Akiyama, K.; Kusajima, M.; Nakashita, H. Regulation of biosynthesis, perception, and functions of strigolactones for promoting arbuscular mycorrhizal symbiosis and managing root parasitic weeds. Pest. Manag. Sci. 2019, 75, 2353–2359. [Google Scholar] [CrossRef]
- Yao, R.; Ming, Z.; Yan, L.; Li, S.; Wang, F.; Ma, S.; Yu, C.; Yang, M.; Chen, L.; Chen, L.; et al. DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 2016, 536, 469–473. [Google Scholar] [CrossRef]
- Hamiaux, C.; Drummond, R.S.M.; Janssen, B.J.; Ledger, S.E.; Cooney, J.M.; Newcomb, R.D.; Snowden, K.C. DAD2 is an α/βhydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol. 2012, 22, 2032–2036. [Google Scholar] [CrossRef] [Green Version]
- de Saint Germain, A.; Clavé, G.; Badet-Denisot, M.-A.; Pillot, J.-P.; Cornu, D.; Le Caer, J.-P.; Burger, M.; Pelissier, F.; Retailleau, P.; Turnbull, C.; et al. An histidine covalent receptor and butenolide complex mediates strigolactone perception. Nat. Chem. Biol. 2016, 12, 787–794. [Google Scholar] [CrossRef] [Green Version]
- Toh, S.; Holbrook-Smith, D.; Stokes, M.E.; Tsuchiya, Y.; McCourt, P. Detection of parasitic plant suicide germination compounds using a high-throughput arabidopsishtl/kai2 strigolactone perception system. Chem. Biol. 2014, 21, 988–998. [Google Scholar] [CrossRef] [Green Version]
- Das, M.; Fernández-Aparicio, M.; Yang, Z.; Huang, K.; Wickett, N.J.; Alford, S.; Wafula, E.K.; DePamphilis, C.; Bouwmeester, H.; Timko, M.P.; et al. Parasitic plants striga and phelipanche dependent upon exogenous strigolactones for germination have retained genes for strigolactone biosynthesis. Am. J. Plant. Sci. 2015, 6, 1151–1166. [Google Scholar] [CrossRef] [Green Version]
- Toh, S.; Holbrook-Smith, D.; Stogios, P.J.; Onopriyenko, O.; Lumba, S.; Tsuchiya, Y.; Savchenko, A.; McCourt, P. Structure-function analysis identifies highly sensitive strigolactone receptors in Striga. Science 2015, 350, 203–207. [Google Scholar] [CrossRef]
- Conn, C.E.; Bythell-Douglas, R.; Neumann, D.; Yoshida, S.; Whittington, B.; Westwood, J.H.; Shirasu, K.; Bond, C.S.; Dyer, K.A.; Nelson, D.C. Convergent evolution of strigolactone perception enabled host detection in parasitic plants. Science 2015, 349, 540–543. [Google Scholar] [CrossRef] [Green Version]
- Tsuchiya, Y.; Yoshimura, M.; Sato, Y.; Kuwata, K.; Toh, S.; Holbrook-Smith, D.; Zhang, H.; McCourt, P.; Itami, K.; Kinoshita, T.; et al. Probing strigolactone receptors in Striga hermonthica with fluorescence. Science 2015, 349, 864–868. [Google Scholar] [CrossRef] [Green Version]
- Yao, R.; Wang, F.; Ming, Z.; Du, X.; Chen, L.; Wang, Y.; Zhang, W.; Deng, H.; Xie, D. ShHTL7 is a non-canonical receptor for strigolactones in root parasitic weeds. Cell Res. 2017, 27, 838–841. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Miyakawa, T.; Nosaki, S.; Nakamura, A.; Lyu, Y.; Nakamura, H.; Ohto, U.; Ishida, H.; Shimizu, T.; Asami, T.; et al. Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga. Nat. Commun. 2018, 9, 1–11. [Google Scholar] [CrossRef]
- Rodríguez-Ojeda, M.I.; Rocío Pineda-Martos, L.C.; Alonso, J.; Fernández-Martínez, J.M.; Pérez-Vich, B.; Velasco, L. A dominant avirulence gene in Orobanche cumana triggers Or5 resistance in sunflower. Weed Res. 2013, 53, 322–327. [Google Scholar]
- Calderón-González, Á.; Pouilly, N.; Muños, S.; Grand, X.; Coque, M.; Velasco, L.; Pérez-Vich, B. A SSR-SNP linkage map of the parasitic weed Orobanche cumana Wallr. including a gene for plant pigmentation. Front. Plant. Sci. 2019, 10, 797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gouzy, J.; Pouilly, N.; Hu, L.; Delavault, P.; Simier, P.; Boniface, M.C.; Louarn, J.; Catrice, O.; Carrère, S.; Cottret, L.; et al. The whole genome sequence of the obligate root parasitic plant Orobanche cumana (sunflower broomrape). In Proceedings of the Plant and Animal Genome XXVII Conference (PAG), San Diego, CA, USA, 12–16 January 2019. [Google Scholar]
- Vranceanu, A.V.; Tudor, V.A.; Stoenescu, F.M.; Pirvu, N. Virulence groups of Orobanche cumana Wallr. [root parasite], differential hosts and resistance sources and genes in sunflower. In Proceedings of the 9th International Conference of Sunflower, Malaga, Spain, 8–13 June 1980. [Google Scholar]
- Pérez-Vich, B.; Akhtouch, B.; Munoz-Ruz, J.; Fernandez-Martinez, J.M.; Jan, C.C. Inheritance of resistance to a highly virulent race F of Orobanche cumana Wallr. in a sunflower line derived from interspecific amphiploids. Helia 2002, 25, 137–143. [Google Scholar] [CrossRef] [Green Version]
- Akhtouch, B.; del Moral, L.; Leon, A.; Velasco, L.; Fernández-Martínez, J.M.; Pérez-Vich, B. Genetic study of recessive broomrape resistance in sunflower. Euphytica 2016, 209, 419–428. [Google Scholar] [CrossRef]
- Velasco, L.; Pérez-Vich, B.; Jan, C.C.; Fernández-Martínez, J.M. Inheritance of resistance to broomrape (Orobanche cumana Wallr.) race F in a sunflower line derived from wild sunflower species. Plant. Breed. 2007, 126, 67–71. [Google Scholar] [CrossRef]
- Velasco, L.; Pérez-Vich, B.; Yassein, A.A.M.; Jan, C.C.; Fernández-Martínez, J.M. Inheritance of resistance to sunflower broomrape (Orobanche cumana Wallr.) in an interspecific cross between Helianthus annuus and Helianthus debilis subsp. tardiflorus. Plant. Breed. 2012, 131, 220–221. [Google Scholar] [CrossRef] [Green Version]
- Imerovski, I.; Dimitrijević, A.; Miladinović, D.; Dedić, B.; Jocić, S.; Tubić, N.K.; Cvejić, S. Mapping of a new gene for resistance to broomrape races higher than F. Euphytica 2016, 209, 281–289. [Google Scholar] [CrossRef]
- Hélène, B.; Gouzy, J.; Grassa, C.J.; Murat, F.; Staton, E.; Cottret, L. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 2017, 7656, 148–152. [Google Scholar]
- Imerovski, I.; Dedić, B.; Cvejić, S.; Miladinović, D.; Jocić, S.; Owens, G.L.; Tubić, N.K.; Rieseberg, L.H. BSA-seq mapping reveals major QTL for broomrape resistance in four sunflower lines. Mol. Breed. 2019, 39, 41. [Google Scholar] [CrossRef]
- Caplan, J.; Padmanabhan, M.; Dinesh-Kumar, S.P. Plant NB-LRR immune receptors: From recognition to transcriptional reprogramming. Cell Host Microbe. 2008, 3, 126–135. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.H.; Qi, D.; Ashfield, T.; Helm, M.; Innes, R.W. Using decoys to expand the recognition specificity of a plant disease resistance protein. Science 2016, 351, 684–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lane, J.A.; Moore, T.H.M.; Child, D.V.; Cardwell, K.F.; Singh, B.B.; Bailey, J.A. Virulence characteristics of a new race of the parasitic angiosperm, Striga gesnerioides, from southern Benin on cowpea (Vigna unguiculata). Euphytica 1994, 72, 183–188. [Google Scholar] [CrossRef]
- Atokple, I.D.K.; Singh, B.B.; Emechebe, A.M. Independent inheritance of Striga and Alectra resistance in cowpea genotype B301. Crop. Sci. 1993, 33, 714–715. [Google Scholar] [CrossRef]
- Singh, B.B.; Emechebe, A.M. Inheritance of Striga resistance in cowpea genotype B301. Crop. Sci. 1990, 30, 879–881. [Google Scholar] [CrossRef]
- Atokple, I.D.K.; Singh, B.B.; Emechebe, A.M. Genetics of resistance to Striga and Alectra in cowpea. J. Hered. 1995, 86, 45–49. [Google Scholar] [CrossRef]
- Ouédraogo, J.T.; Maheshwari, V.; Berner, D.K.; St-Pierre, C.-A.; Belzile, F.; Timko, M.P. Identification of AFLP markers linked to resistance of cowpea (Vigna unguiculata L.) to parasitism by Striga gesnerioides. Theor. Appl. Genet. 2001, 102, 1029–1036. [Google Scholar] [CrossRef]
- Li, J.; Lis, K.E.; Timko, M.P. Molecular genetics of race-specific resistance of cowpea to Striga gesnerioides (Willd.). Pest. Manag. Sci. 2009, 65, 520–527. [Google Scholar] [CrossRef]
- Timko, M.P.; Rushton, P.J.; Laudeman, T.W.; Bokowiec, M.T.; Chipumuro, E.; Cheung, F.; Town, C.D.; Chen, X. Sequencing and analysis of the gene-rich space of cowpea. BMC Genomics 2008, 9, 103. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Timko, M.P. Gene-for-gene resistance in Striga-cowpea associations. Science 2009, 325, 1094. [Google Scholar] [CrossRef]
- Yang, Z.; Wafula, E.K.; Honaas, L.A.; Zhang, H.; Das, M.; Fernandez-Aparicio, M.; Huang, K.; Bandaranayake, P.C.G.; Wu, B.; Der, J.P.; et al. Comparative transcriptome analyses reveal core parasitism genes and suggest gene duplication and repurposing as sources of structural novelty. Mol. Biol. Evol. 2015, 32, 767–790. [Google Scholar] [CrossRef] [Green Version]
- Su, C.; Liu, H.; Wafula, E.K.; Honaas, L.; Pamphilis, C.W.; Timko, M.P. SHR4z, a novel decoy effector from the haustorium of the parasitic weed Striga gesnerioides, suppresses host plant immunity. New Phytol. 2020, 226, 891–908. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Qi, J.; Yue, J.; Huang, J.; Sun, T.; Li, S.; Wen, J.-F.; Hettenhausen, C.; Wu, J.; Wang, L.; et al. Root parasitic plant Orobanche aegyptiaca and shoot parasitic plant Cuscuta australis obtained Brassicaceae-specific strictosidine synthase-like genes by horizontal gene transfer. BMC Plant. Biol. 2014, 14, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Hegenauer, V.; Fürst, U.; Kaiser, B.; Smoker, M.; Zipfel, C.; Felix, G.; Stahl, M.; Albert, M. Detection of the plant parasite Cuscuta reflexa by a tomato cell surface receptor. Science 2016, 353, 478–481. [Google Scholar] [CrossRef]
- Bradley, J.; Qiu, S.; Butlin, R.; Chaudhuri, R.; Scholes, J. The identification of candidate pathogenicity-related genes from the genome of Striga hermonthica. In Proceedings of the 15th World Congress on Parasitic Plants, Amsterdam, The Netherlands, 30 June–5 July 2019. [Google Scholar]
- Wang, Y.; Xu, Y.; Sun, Y.; Wang, H.; Qi, J.; Wan, B.; Ye, W.; Lin, Y.; Shao, Y.; Dong, S.; et al. Leucine-rich repeat receptor-like gene screen reveals that Nicotiana RXEG1 regulates glycoside hydrolase 12 MAMP detection. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Westwood, J.H.; DePamphilis, C.W.; Das, M.; Fernández-Aparicio, M.; Honaas, L.A.; Timko, M.P.; Wafula, E.K.; Wickett, N.J.; Yoder, J.I. The parasitic plant genome project: New tools for understanding the biology of Orobanche and Striga. Weed Sci. 2012, 60, 295–306. [Google Scholar] [CrossRef]
- Yang, C.; Fu, F.; Zhang, N.; Wang, J.; Hu, L.; Islam, F.; Zhou, W. Transcriptional profiling of underground interaction of two contrasting sunflower cultivars with the root parasitic weed Orobanche cumana. Plant. Soil 2020, 450, 303–321. [Google Scholar] [CrossRef]
- Li, J.; Liu, H.; Yang, C.; Wang, J.; Yan, G.; Si, P.; Bai, Q.; Lu, Z.; Zhou, W.; Xu, L. Genome-wide identification of MYB genes and expression analysis under different biotic and abiotic stresses in Helianthus annuus L. Ind. Crops Prod. 2020, 143, 111924. [Google Scholar] [CrossRef]
- Bellot, S.; Renner, S.S. Exploring new dating approaches for parasites: The worldwide Apodanthaceae (Cucurbitales) as an example. Mol. Phylogenet. Evol. 2014, 80, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Xi, Z.; Wang, Y.; Bradley, R.K.; Sugumaran, M.; Marx, C.J.; Rest, J.S.; Davis, C.C. Massive mitochondrial gene transfer in a parasitic flowering plant clade. PLoS Genet. 2013, 9, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, G.; LeBlanc, M.L.; Wafula, E.K.; DePamphilis, C.W.; Westwood, J.H. Genomic-scale exchange of mRNA between a parasitic plant and its hosts. Science 2014, 345, 808–811. [Google Scholar] [CrossRef]
- Fan, W.; Zhu, A.; Kozaczek, M.; Shah, N.; Pabón-Mora, N.; González, F.; Mower, J.P. Limited mitogenomic degradation in response to a parasitic lifestyle in Orobanchaceae. Sci. Rep. 2016, 6, 36285. [Google Scholar] [CrossRef]
- Vogel, A.; Schwacke, R.; Denton, A.K.; Usadel, B.; Hollmann, J.; Fischer, K.; Bolger, A.; Schmidt, M.H.W.; Bolger, M.E.; Gundlach, H.; et al. Footprints of parasitism in the genome of the parasitic flowering plant Cuscuta campestris. Nat. Commun. 2018, 9, 1–11. [Google Scholar]
- Sun, G.; Xu, Y.; Liu, H.; Sun, T.; Zhang, J.; Hettenhausen, C.; Shen, G.; Qi, J.; Qin, Y.; Li, J.; et al. Large-scale gene losses underlie the genome evolution of parasitic plant Cuscuta australis. Nat. Commun. 2018, 9, 1–8. [Google Scholar]
- Naumann, J.; Der, J.P.; Wafula, E.K.; Jones, S.S.; Wagner, S.T.; Honaas, L.A.; Ralph, P.E.; Bolin, J.F.; Maass, E.; Neinhuis, C.; et al. Detecting and characterizing the highly divergent plastid genome of the nonphotosynthetic parasitic plant Hydnora visseri (Hydnoraceae). Genome Biol. Evol. 2016, 8, 345–363. [Google Scholar]
- Schneider, A.C.; Chun, H.; Stefanović, S.; Baldwin, B.G. Punctuated plastome reduction and host–parasite horizontal gene transfer in the holoparasitic plant genus Aphyllon. Proc. R. Soc. B Biol. Sci. 2018, 285, 1–8. [Google Scholar]
- Yoshida, S.; Kim, S.; Wafula, E.K.; Tanskanen, J.; Kim, Y.M.; Honaas, L.; Yang, Z.; Spallek, T.; Conn, C.E.; Ichihashi, Y.; et al. Genome sequence of Striga asiatica provides insight into the evolution of plant parasitism. Curr. Biol. 2019, 29, 3041–3052. [Google Scholar]
- Yang, Z.; Wafula, E.K.; Kim, G.; Shahid, S.; McNeal, J.R.; Ralph, P.E.; Timilsena, P.R.; Yu, W.B.; Kelly, E.A.; Zhang, H.; et al. Convergent horizontal gene transfer and cross-talk of mobile nucleic acids in parasitic plants. Nat.Plants 2019, 5, 991–1001. [Google Scholar]
- Yang, Z.; Zhang, Y.; Wafula, E.K.; Honaas, L.A.; Ralph, P.E.; Jones, S.; Clarke, C.R.; Liu, S.; Su, C.; Zhang, H.; et al. Horizontal gene transfer is more frequent with increased heterotrophy and contributes to parasite adaptation. Proc. Natl. Acad. Sci. USA 2016, 113, E7010–E7019. [Google Scholar]
- Xi, Z.; Bradley, R.K.; Wurdack, K.J.; Wong, K.M.; Sugumaran, M.; Bomblies, K.; Rest, J.S.; Davis, C.C. Horizontal transfer of expressed genes in a parasitic flowering plant. BMC Genomics 2012, 13, 227. [Google Scholar]
- Bellot, S.; Cusimano, N.; Luo, S.; Sun, G.; Zarre, S.; Gröger, A.; Temsch, E.; Renner, S.S. Assembled plastid and mitochondrial genomes, as well as nuclear genes, place the parasite family Cynomoriaceae in the Saxifragales. Genome Biol. Evol. 2016, 8, 2214–2230. [Google Scholar] [PubMed] [Green Version]
- Lopez, L.; Bellis, E.S.; Wafula, E.; Hearne, S.J.; Honaas, L.; Ralph, P.E.; Timko, M.P.; Unachukwu, N.; DePamphilis, C.W.; Lasky, J.R. Transcriptomics of host-specific interactions in natural populations of the parasitic plant purple witchweed ( Striga hermonthica ). Weed Sci. 2019, 67, 397–411. [Google Scholar]
- Ranjan, A.; Ichihashi, Y.; Farhi, M.; Zumstein, K.; Townsley, B.; David-Schwartz, R.; Sinha, N.R. De novo assembly and characterization of the transcriptome of the parasitic weed dodder identifies genes associated with plant parasitism. Plant. Physiol. 2014, 166, 1186–1199. [Google Scholar]
- Kennan, R.M.; Wong, W.; Dhungyel, O.P.; Han, X.; Wong, D.; Parker, D.; Rosado, C.J.; Law, R.H.P.; McGowan, S.; Reeve, S.B.; et al. The subtilisin-like protease Aprv2 is required for virulence and uses a novel disulphide-tethered exosite to bind substrates. PLoS Pathog. 2010, 6, e1001210. [Google Scholar]
- Antão, C.M.; Malcata, F.X. Plant serine proteases: Biochemical, physiological and molecular features. Plant. Physiol. Biochem. 2005, 43, 637–650. [Google Scholar]
- Moffett, P. Mechanisms of recognition in dominant R gene mediated resistance. Adv. Virus Res. 2009, 75, 1–229. [Google Scholar] [PubMed]
- Adachi, H.; Derevnina, L.; Kamoun, S. NLR singletons, pairs, and networks: Evolution, assembly, and regulation of the intracellular immunoreceptor circuitry of plants. Curr. Opin. Plant. Biol. 2019, 50, 121–131. [Google Scholar] [PubMed] [Green Version]
- Wirthmueller, L.; Maqbool, A.; Banfield, M.J. On the front line: Structural insights into plant–pathogen interactions. Nat. Rev. Microbiol. 2013, 11, 761–776. [Google Scholar] [PubMed]
- McHale, L.; Tan, X.; Koehl, P.; Michelmore, R.W. Plant NBS-LRR proteins: Adaptable guards. Genome Biol. 2006, 7, 1–11. [Google Scholar]
- Koehl, P.; Delarue, M. A self consistent mean field approach to simultaneous gap closure and side-chain positioning in homology modelling. Nat. Struct. Mol. Biol. 1995, 2, 163–170. [Google Scholar]
- Steele, J.F.C.; Hughes, R.K.; Banfield, M.J. Structural and biochemical studies of an NB-ARC domain from a plant NLR immune receptor. PLoS ONE 2019, 14, e0221226. [Google Scholar]
- Holt, B.F.; Boyes, D.C.; Ellerström, M.; Siefers, N.; Wiig, A.; Kauffman, S.; Grant, M.R.; Dangl, J.L. An evolutionarily conserved mediator of plant disease resistance gene function is required for normal Arabidopsis development. Dev. Cell 2002, 2, 807–817. [Google Scholar]
- Machens, F.; Becker, M.; Umrath, F.; Hehl, R. Identification of a novel type of WRKY transcription factor binding site in elicitor-responsive cis-sequences from Arabidopsis thaliana. Plant. Mol. Biol. 2014, 84, 371–385. [Google Scholar] [PubMed]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant. Sci. 2010, 15, 247–258. [Google Scholar] [PubMed]
- Yang, Y.; Zhou, Y.; Chi, Y.; Fan, B.; Chen, Z. Characterization of soybean WRKY gene family and identification of soybean WRKY genes that promote resistance to soybean cyst nematode. Sci. Rep. 2017, 7, 17804. [Google Scholar] [PubMed] [Green Version]
- Deslandes, L.; Olivier, J.; Theulieres, F.; Hirsch, J.; Feng, D.X.; Bittner-Eddy, P.; Beynon, J.; Marco, Y. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc. Natl. Acad. Sci. USA 2002, 99, 2404–2409. [Google Scholar] [PubMed] [Green Version]
- Keebaugh, E.S.; Schlenke, T.A. Insights from natural host–parasite interactions: The Drosophila model. Dev. Comp. Immunol. 2014, 42, 111–123. [Google Scholar]
- Dodds, P.N.; Lawrence, G.J.; Catanzariti, A.-M.; Teh, T.; Wang, C.-I.A.; Ayliffe, M.A.; Kobe, B.; Ellis, J.G. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. USA 2006, 103, 8888–8893. [Google Scholar]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar]
- DeYoung, B.J.; Innes, R.W. Plant NBS-LRR proteins in pathogen sensing and host defense. Nat. Immunol. 2006, 7, 1243–1249. [Google Scholar]
- Chisholm, S.T.; Coaker, G.; Day, B.; Staskawicz, B.J. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 2006, 124, 803–814. [Google Scholar]
- Upson, J.L.; Zess, E.K.; Białas, A.; Wu, C.; Kamoun, S. The coming of age of EvoMPMI: Evolutionary molecular plant–microbe interactions across multiple timescales. Curr. Opin. Plant. Biol. 2018, 44, 108–116. [Google Scholar]
- Runo, S.; Kuria, E.K. Habits of a highly successful cereal killer, Striga. PLoS Pathog. 2018, 14, e1006731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bellis, E.S.; Kelly, E.A.; Lorts, C.M.; Gao, H.; DeLeo, V.L.; Rouhan, G.; Budden, A.; Bhaskara, G.B.; Hu, Z.; Muscarella, R.; et al. Genomics of sorghum local adaptation to a parasitic plant. Proc. Natl. Acad. Sci. USA 2020, 117, 4243–4251. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Stam, R.; Cano, L.M.; Song, J.; Sklenar, J.; Yoshida, K.; Bozkurt, T.O.; Oliva, R.; Liu, Z.; Tian, M.; et al. Effector specialization in a lineage of the Irish potato famine pathogen. Science 2014, 343, 552–555. [Google Scholar] [CrossRef] [PubMed]
- Białas, A.; Zess, E.K.; De la Concepcion, J.C.; Franceschetti, M.; Pennington, H.G.; Yoshida, K.; Upson, J.L.; Chanclud, E.; Wu, C.-H.; Langner, T.; et al. Lessons in effector and NLR biology of plant-microbe systems. Mol. Plant. Microbe. Interact. 2018, 31, 34–45. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, K.; Saitoh, H.; Fujisawa, S.; Kanzaki, H.; Matsumura, H.; Yoshida, K.; Tosa, Y.; Chuma, I.; Takano, Y.; Win, J.; et al. Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae. Plant. Cell 2009, 21, 1573–1591. [Google Scholar] [CrossRef] [Green Version]
- Maqbool, A.; Saitoh, H.; Franceschetti, M.; Stevenson, C.E.M.; Uemura, A.; Kanzaki, H.; Kamoun, S.; Terauchi, R.; Banfield, M.J. Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor. Elife 2015, 4. [Google Scholar]
- Fouché, S.; Plissonneau, C.; Croll, D. The birth and death of effectors in rapidly evolving filamentous pathogen genomes. Curr. Opin. Microbiol. 2018, 46, 34–42. [Google Scholar] [CrossRef]
- Dong, S.; Raffaele, S.; Kamoun, S. The two-speed genomes of filamentous pathogens: Waltz with plants. Curr. Opin. Genet. Dev. 2015, 35, 57–65. [Google Scholar] [CrossRef]
- Dong, Y.; Li, Y.; Zhao, M.; Jing, M.; Liu, X.; Liu, M.; Guo, X.; Zhang, X.; Chen, Y.; Liu, Y.; et al. Global genome and transcriptome analyses of Magnaporthe oryzae epidemic isolate 98-06 uncover novel effectors and pathogenicity-related genes, revealing gene gain and lose dynamics in genome evolution. PLoS Pathog. 2015, 11, e1004801. [Google Scholar]
- Raffaele, S.; Kamoun, S. Genome evolution in filamentous plant pathogens: Why bigger can be better. Nat. Rev. Microbiol. 2012, 10, 417–430. [Google Scholar] [CrossRef]
- Hartmann, F.E.; Croll, D. Distinct trajectories of massive recent gene gains and losses in populations of a microbial eukaryotic pathogen. Mol. Biol. Evol. 2017, 34, 2808–2822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Möller, M.; Stukenbrock, E.H. Evolution and genome architecture in fungal plant pathogens. Nat. Rev. Microbiol. 2017, 15, 756–771. [Google Scholar] [PubMed]
- Brown, J.K.M.; Tellier, A. Plant-parasite coevolution: Bridging the gap between genetics and ecology. Annu. Rev. Phytopathol. 2011, 49, 345–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricroch, A.; Clairand, P.; Harwood, W. Use of CRISPR systems in plant genome editing: Toward new opportunities in agriculture. Emerg. Top. Life Sci. 2017, 1, 169–182. [Google Scholar]
- Butt, H.; Jamil, M.; Wang, J.Y.; Al-Babili, S.; Mahfouz, M. Engineering plant architecture via CRISPR/Cas9-mediated alteration of strigolactone biosynthesis. BMC Plant. Biol. 2018, 18, 174. [Google Scholar] [CrossRef] [Green Version]
- Bari, V.K.; Nassar, J.A.; Kheredin, S.M.; Gal-On, A.; Ron, M.; Britt, A.; Steele, D.; Yoder, J.; Aly, R. CRISPR/Cas9-mediated mutagenesis of CAROTENOID CLEAVAGE DIOXYGENASE 8 in tomato provides resistance against the parasitic weed Phelipanche aegyptiaca. Sci. Rep. 2019, 9, 11438. [Google Scholar] [CrossRef]
- Cheng, X.; Floková, K.; Bouwmeester, H.; Ruyter-Spira, C. The role of endogenous strigolactones and their interaction with aba during the infection process of the parasitic weed phelipanche ramosa in tomato plan. Front. Plant. Sci. 2017, 8, 392. [Google Scholar] [CrossRef] [Green Version]
- Bennett, T.; Sieberer, T.; Willett, B.; Booker, J.; Luschnig, C.; Leyser, O. The arabidopsis Max pathway controls shoot branching by regulating auxin transport. Curr. Biol. 2006, 16, 553–563. [Google Scholar] [CrossRef] [Green Version]
- Harvey, J.J.W.; Lewsey, M.G.; Patel, K.; Westwood, J.; Heimstadt, S.; Carr, J.P.; Baulcombe, D.C. An antiviral defense role of AGO2 in plants. PLoS ONE 2011, 6, e14639. [Google Scholar] [CrossRef]
- Baulcombe, D. RNA silencing in plants. Nature 2004, 431, 356–363. [Google Scholar]
- Qi, T.; Guo, J.; Peng, H.; Liu, P.; Kang, Z.; Guo, J. Host-induced gene silencing: A powerful strategy to control diseases of wheat and barley. Int. J. Mol. Sci. 2019, 20, 206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, S.-W. RNA-based antiviral immunity. Nat. Rev. Immunol. 2010, 10, 632–644. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Allen, R.; Davis, E.L.; Baum, T.J.; Hussey, R.S. Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Natl. Acad. Sci. 2006, 103, 14302–14306. [Google Scholar] [CrossRef] [Green Version]
- Tomilov, A.A.; Tomilova, N.B.; Wroblewski, T.; Michelmore, R.; Yoder, J.I. Trans-specific gene silencing between host and parasitic plants. Plant. J. 2008, 56, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Westwood, J.H.; Roney, J.K.; Khatibi, P.A.; Stromberg, V.K. RNA translocation between parasitic plants and their hosts. Pest. Manag. Sci. 2009, 65, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Alakonya, A.; Kumar, R.; Koenig, D.; Kimura, S.; Townsley, B.; Runo, S.; Garces, H.M.; Kang, J.; Yanez, A.; David-Schwartz, R.; et al. Interspecific RNA interference of SHOOT MERISTEMLESS-like disrupts Cuscuta pentagona plant parasitism. Plant. Cell 2012, 24, 3153–3166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirigia, D.; Runo, S.; Alakonya, A. A virus-induced gene silencing (VIGS) system for functional genomics in the parasitic plant Striga hermonthica. Plant. Methods 2014, 10, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Framond, A.; Rich, P.J.; McMillan, J.; Ejeta, G. Effects on Striga parasitism of transgenic maize armed with RNAi constructs targeting essential S. asiatica genes. In Integrating New Technologies for Striga Control; Gebisa, E., Jonathan, G., Eds.; World Scientific: Singapore, 2007; pp. 185–196. [Google Scholar]
- Aly, R.; Cholakh, H.; Joel, D.M.; Leibman, D.; Steinitz, B.; Zelcer, A.; Naglis, A.; Yarden, O.; Gal-On, A. Gene silencing of mannose 6-phosphate reductase in the parasitic weed Orobanche aegyptiaca through the production of homologous dsRNA sequences in the host plant. Plant. Biotechnol. J. 2009, 7, 487–498. [Google Scholar] [CrossRef]
- Dubey, N.K.; Eizenberg, H.; Leibman, D.; Wolf, D.; Edelstein, M.; Abu-Nassar, J.; Marzouk, S.; Gal-On, A.; Aly, R. Enhanced host-parasite resistance based on down-regulation of Phelipanche aegyptiaca target genes is likely by mobile small RNA. Front. Plant. Sci. 2017, 8, 1574. [Google Scholar] [CrossRef] [Green Version]
- Kohlen, W.; Charnikhova, T.; Lammers, M.; Pollina, T.; Tóth, P.; Haider, I.; Pozo, M.J.; Maagd, R.A.; Ruyter-Spira, C.; Bouwmeester, H.J.; et al. The tomato CAROTENOID CLEAVAGE DIOXYGENASE 8 (SlCCD8) regulates rhizosphere signaling, plant architecture and affects reproductive development through strigolactone biosynthesis. New Phytol. 2012, 196, 535–547. [Google Scholar] [CrossRef]
- Waldie, T.; McCulloch, H.; Leyser, O. Strigolactones and the control of plant development: Lessons from shoot branching. Plant. J. 2014, 79, 607–622. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, K.P.; Vishwakarma, C.; Sahoo, S.P.; Lima, J.M.; Nath, M.; Dokku, P.; Gacche, R.N.; Mohapatra, T.; Robin, S.; Sarla, N.; et al. A substitution mutation in OsCCD7 cosegregates with dwarf and increased tillering phenotype in rice. J. Genet. 2014, 93, 389–401. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Chen, L.; He, J.; Yu, W. Knocking out of carotenoid catabolic genes in rice fails to boost carotenoid accumulation, but reveals a mutation in strigolactone biosynthesis. Plant. Cell Rep. 2017, 36, 1533–1545. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Zhang, T.; Xu, B.; Jia, L.; Xiao, B.; Liu, H.; Liu, L.; Yan, H.; Xia, Q. Crispr/cas9-mediated mutagenesis of carotenoid cleavage dioxygenase 8 (ccd8) in tobacco affects shoot and root architecture. Int. J. Mol. Sci. 2018, 19, 1062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ledger, S.E.; Janssen, B.J.; Karunairetnam, S.; Wang, T.; Snowden, K.C. Modified CAROTENOID CLEAVAGE DIOXYGENASE8 expression correlates with altered branching in kiwifruit (Actinidia chinensis). New Phytol. 2010, 188, 803–813. [Google Scholar] [CrossRef]
- Tsuchiya, Y.; Yoshimura, M.; Hagihara, S. The dynamics of strigolactone perception in Striga hermonthica: A working hypothesis. J. Exp. Bot. 2018, 69, 2281–2290. [Google Scholar] [CrossRef] [Green Version]
- Vogel, J.T.; Walter, M.H.; Giavalisco, P.; Lytovchenko, A.; Kohlen, W.; Charnikhova, T.; Simkin, A.J.; Goulet, C.; Strack, D.; Bouwmeester, H.J.; et al. SlCCD7 controls strigolactone biosynthesis, shoot branching and mycorrhiza-induced apocarotenoid formation in tomato. Plant. J. 2010, 61, 300–311. [Google Scholar] [CrossRef]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, L.; Wang, J.; Yang, C.; Islam, F.; Bouwmeester, H.J.; Muños, S.; Zhou, W. The Effect of Virulence and Resistance Mechanisms on the Interactions between Parasitic Plants and Their Hosts. Int. J. Mol. Sci. 2020, 21, 9013. https://doi.org/10.3390/ijms21239013
Hu L, Wang J, Yang C, Islam F, Bouwmeester HJ, Muños S, Zhou W. The Effect of Virulence and Resistance Mechanisms on the Interactions between Parasitic Plants and Their Hosts. International Journal of Molecular Sciences. 2020; 21(23):9013. https://doi.org/10.3390/ijms21239013
Chicago/Turabian StyleHu, Luyang, Jiansu Wang, Chong Yang, Faisal Islam, Harro J. Bouwmeester, Stéphane Muños, and Weijun Zhou. 2020. "The Effect of Virulence and Resistance Mechanisms on the Interactions between Parasitic Plants and Their Hosts" International Journal of Molecular Sciences 21, no. 23: 9013. https://doi.org/10.3390/ijms21239013