A Network-Based Approach to Explore the Mechanisms of Uncaria Alkaloids in Treating Hypertension and Alleviating Alzheimer’s Disease
Abstract
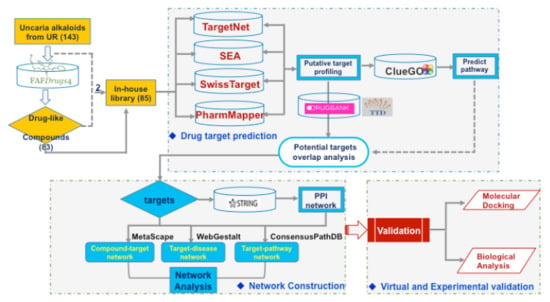
:1. Introduction
2. Results
2.1. The In-House Library of Uncaria Alkaloids and Drug-Likeness Screening
2.2. Putative Target Prediction for the Candidate Targets of Uncaria Alkaloids
2.3. Identification of Approved Druggable Targets in AD and Hypertension
2.4. Pathways in Compound-Target Network
2.5. Network Construction, Topological Analysis, and GO/KEGG Function and Pathway Analysis
2.6. Pathways in Target-Disease Network
2.7. Pathways in Target-Pathway Network
2.8. Integrative Network Analysis and Target Selection
2.9. Molecular Docking and Biological Validation
3. Discussion
4. Materials and Methods
4.1. Compound Library Construction and Drug-Likeness Screening
4.2. Prediction of Uncaria Alkaloid Targets and Their Overall Biological Functions
4.3. Identification of Druggable Alzheimer’s Disease (AD) and Hypertension (HTN) Associated Disease Targets
4.4. Network Construction, Topological Analysis and GO/KEGG Function Analysis
4.5. Molecular Docking
4.6. In Vitro Butyrylcholinesterase (BChE) Enzyme Activity Assays
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s disease |
| BChE | butyrylcholinesterase |
| HTN | hypertension |
| PPI | protein–protein interaction |
| TCM | Traditional Chinese Medicine |
References
- Zhang, Q.; Zhao, J.J.; Xu, J.; Feng, F.; Qu, W. Medicinal uses, phytochemistry and pharmacology of the genus Uncaria. J. Ethnopharmacol. 2015, 173, 48–80. [Google Scholar] [CrossRef] [PubMed]
- Laus, G. Advances in chemistry and bioactivity of the genus Uncaria. Phytother. Res. 2004, 18, 259–274. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhao, Y.H.; Zeng, C.Q.; Zeng, Y. Research progress in pharmacological effects of Uncaria Hook on Alzheimer disease models. Yao Xue Xue Bao 2016, 51, 536–542. [Google Scholar] [PubMed]
- Feng, Z.; Hou, J.; Yu, Y.; Wu, W.; Deng, Y.; Wang, X.; Zhi, H.; Zhang, L.; Wu, W.; Guo, D.A. Dissecting the Metabolic Phenotype of the Antihypertensive Effects of Five Uncaria Species on Spontaneously Hypertensive Rats. Front. Pharmacol. 2019, 10, 845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ndagijimana, A.; Wang, X.; Pan, G.; Zhang, F.; Feng, H.; Olaleye, O. A review on indole alkaloids isolated from Uncaria rhynchophylla and their pharmacological studies. Fitoterapia 2013, 86, 35–47. [Google Scholar] [CrossRef]
- Fujiwara, H.; Iwasaki, K.; Furukawa, K.; Seki, T.; He, M.; Maruyama, M.; Tomita, N.; Kudo, Y.; Higuchi, M.; Saido, T.C.; et al. Uncaria rhynchophylla, a Chinese medicinal herb, has potent antiaggregation effects on Alzheimer’s beta-amyloid proteins. J. Neurosci. Res. 2006, 84, 427–433. [Google Scholar] [CrossRef]
- Yang, W.; Ip, S.P.; Liu, L.; Xian, Y.F.; Lin, Z.X. Uncaria rhynchophylla and Its Major Constituents on Central Nervous System: A Review on Their Pharmacological Actions. Curr. Vasc. Pharmacol. 2019. [Google Scholar] [CrossRef]
- Shao, H.; Mi, Z.; Ji, W.G.; Zhang, C.H.; Zhang, T.; Ren, S.C.; Zhu, Z.R. Rhynchophylline Protects Against the Amyloid beta-Induced Increase of Spontaneous Discharges in the Hippocampal CA1 Region of Rats. Neurochem. Res. 2015, 40, 2365–2373. [Google Scholar] [CrossRef]
- Shin, S.J.; Jeong, Y.; Jeon, S.G.; Kim, S.; Lee, S.K.; Choi, H.S.; Im, C.S.; Kim, S.H.; Kim, S.H.; Park, J.H.; et al. Uncaria rhynchophylla ameliorates amyloid beta deposition and amyloid beta-mediated pathology in 5XFAD mice. Neurochem. Int. 2018, 121, 114–124. [Google Scholar] [CrossRef]
- Yang, Y.; Ji, W.G.; Zhu, Z.R.; Wu, Y.L.; Zhang, Z.Y.; Qu, S.C. Rhynchophylline suppresses soluble Abeta1-42-induced impairment of spatial cognition function via inhibiting excessive activation of extrasynaptic NR2B-containing NMDA receptors. Neuropharmacology 2018, 135, 100–112. [Google Scholar] [CrossRef]
- Cao, J.; Hou, J.; Ping, J.; Cai, D. Advances in developing novel therapeutic strategies for Alzheimer’s disease. Mol. Neurodegener. 2018, 13, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pardridge, W.M. Alzheimer’s disease: Future drug development and the blood-brain barrier. Expert Opin. Investig. Drugs 2019, 28, 569–572. [Google Scholar] [CrossRef] [PubMed]
- Sala Frigerio, C.; De Strooper, B. Alzheimer’s Disease Mechanisms and Emerging Roads to Novel Therapeutics. Annu. Rev. Neurosci. 2016, 39, 57–79. [Google Scholar] [CrossRef] [PubMed]
- Carnevale, D.; Perrotta, M.; Lembo, G.; Trimarco, B. Pathophysiological Links among Hypertension and Alzheimer’s Disease. High Blood Press. Cardiovasc. Prev. 2016, 23, 3–7. [Google Scholar] [CrossRef] [Green Version]
- Csiszar, A.; Tucsek, Z.; Toth, P.; Sosnowska, D.; Gautam, T.; Koller, A.; Deak, F.; Sonntag, W.E.; Ungvari, Z. Synergistic effects of hypertension and aging on cognitive function and hippocampal expression of genes involved in beta-amyloid generation and Alzheimer’s disease. Am. J. Physiol. Heart Circ. Physiol. 2013, 305, H1120–H1130. [Google Scholar] [CrossRef] [Green Version]
- Jeon, S.Y.; Byun, M.S.; Yi, D.; Lee, J.H.; Choe, Y.M.; Ko, K.; Sohn, B.K.; Choi, H.J.; Lee, J.Y.; Lee, D.Y.; et al. Influence of hypertension on brain amyloid deposition and Alzheimer’s disease signature neurodegeneration. Neurobiol. Aging 2019, 75, 62–70. [Google Scholar] [CrossRef]
- Sundar, U.; Manwatkar, A.A.; Joshi, A.R.; Bhandarkar, P. The Effect of Hypertension and Diabetes Mellitus on White Matter Changes in MRI Brain: A Comparative Study between Patients with Alzheimer’s Disease and an Age-matched Control Group. J. Assoc. Physicians India 2019, 67, 14–17. [Google Scholar]
- Shih, Y.H.; Wu, S.Y.; Yu, M.; Huang, S.H.; Lee, C.W.; Jiang, M.J.; Lin, P.Y.; Yang, T.T.; Kuo, Y.M. Hypertension Accelerates Alzheimer’s Disease-Related Pathologies in Pigs and 3xTg Mice. Front. Aging Neurosci. 2018, 10, 73. [Google Scholar] [CrossRef] [Green Version]
- Wiesmann, M.; Roelofs, M.; van der Lugt, R.; Heerschap, A.; Kiliaan, A.J.; Claassen, J.A. Angiotensin II, hypertension and angiotensin II receptor antagonism: Roles in the behavioural and brain pathology of a mouse model of Alzheimer’s disease. J. Cereb. Blood Flow Metab. 2017, 37, 2396–2413. [Google Scholar] [CrossRef]
- Kruyer, A.; Soplop, N.; Strickland, S.; Norris, E.H. Chronic Hypertension Leads to Neurodegeneration in the TgSwDI Mouse Model of Alzheimer’s Disease. Hypertension 2015, 66, 175–182. [Google Scholar] [CrossRef] [Green Version]
- Israeli-Korn, S.D.; Masarwa, M.; Schechtman, E.; Abuful, A.; Strugatsky, R.; Avni, S.; Farrer, L.A.; Friedland, R.P.; Inzelberg, R. Hypertension increases the probability of Alzheimer’s disease and of mild cognitive impairment in an Arab community in northern Israel. Neuroepidemiology 2010, 34, 99–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.H.; Roe, C.M.; Morris, J.C. Relationship between late-life hypertension, blood pressure, and Alzheimer’s disease. Am. J. Alzheimer’s Dis. Other Dement. 2011, 26, 457–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cifuentes, D.; Poittevin, M.; Dere, E.; Broqueres-You, D.; Bonnin, P.; Benessiano, J.; Pocard, M.; Mariani, J.; Kubis, N.; Merkulova-Rainon, T.; et al. Hypertension accelerates the progression of Alzheimer-like pathology in a mouse model of the disease. Hypertension 2015, 65, 218–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, T.; Hanyu, H.; Akai, T.; Takasaki, A.; Sakurai, H.; Iwamoto, T. A patient with early Alzheimer’s disease who showed improvement of cognitive function and cerebral perfusion by combined therapy of nilvadipine and PPAR gamma agonists. Nihon Ronen Igakkai Zasshi 2008, 45, 428–433. [Google Scholar] [CrossRef] [Green Version]
- Lawlor, B.; Kennelly, S.; O’Dwyer, S.; Cregg, F.; Walsh, C.; Coen, R.; Kenny, R.A.; Howard, R.; Murphy, C.; Adams, J.; et al. NILVAD protocol: A European multicentre double-blind placebo-controlled trial of nilvadipine in mild-to-moderate Alzheimer’s disease. BMJ Open 2014, 4, e006364. [Google Scholar] [CrossRef] [PubMed]
- Meulenbroek, O.; O’Dwyer, S.; de Jong, D.; van Spijker, G.; Kennelly, S.; Cregg, F.; Olde Rikkert, M.; Abdullah, L.; Wallin, A.; Walsh, C.; et al. European multicentre double-blind placebo-controlled trial of Nilvadipine in mild-to-moderate Alzheimer’s disease-the substudy protocols: NILVAD frailty; NILVAD blood and genetic biomarkers; NILVAD cerebrospinal fluid biomarkers; NILVAD cerebral blood flow. BMJ Open 2016, 6, e011584. [Google Scholar] [CrossRef] [Green Version]
- De Jong, D.L.K.; de Heus, R.A.A.; Rijpma, A.; Donders, R.; Olde Rikkert, M.G.M.; Gunther, M.; Lawlor, B.A.; van Osch, M.J.P.; Claassen, J. Effects of Nilvadipine on Cerebral Blood Flow in Patients with Alzheimer Disease. Hypertension 2019, 74, 413–420. [Google Scholar] [CrossRef]
- Tu, Y. Artemisinin-A Gift from Traditional Chinese Medicine to the World (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 2016, 55, 10210–10226. [Google Scholar] [CrossRef]
- Kong, L.Y.; Tan, R.X. Artemisinin, a miracle of traditional Chinese medicine. Nat. Prod. Rep. 2015, 32, 1617–1621. [Google Scholar] [CrossRef]
- Li, S. Advances in TCM symptomatology of rheumatoid arthritis. J. Tradit. Chin. Med. 2002, 22, 137–142. [Google Scholar]
- Li, S.; Zhang, B. Traditional Chinese medicine network pharmacology: Theory, methodology and application. Chin. J. Nat. Med. 2013, 11, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Huai, Y.; Miao, Z.; Qian, A.; Wang, Y. Systems Pharmacology for Investigation of the Mechanisms of Action of Traditional Chinese Medicine in Drug Discovery. Front. Pharmacol. 2019, 10, 743. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Bing, Z.; Bi, Z.; Liu, M.; Lu, T.; Xun, Y.; Wei, Z.; Yang, K. Top-100 Most Cited Publications Concerning Network Pharmacology: A Bibliometric Analysis. Evid. Based Complement. Alternat. Med. 2019, 2019, 1704816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, J.; Wu, M.; Wang, H.; Li, S.; Wang, X.; Li, Y.; Wang, D.; Li, S. Network Pharmacology to Unveil the Biological Basis of Health-Strengthening Herbal Medicine in Cancer Treatment. Cancers 2018, 10, 461. [Google Scholar] [CrossRef] [Green Version]
- Li, J.Y.; Chen, H.Y.; Dai, W.J.; Lv, Q.J.; Chen, C.Y. Artificial Intelligence Approach to Investigate the Longevity Drug. J. Phys. Chem. Lett. 2019, 4947–4961. [Google Scholar] [CrossRef]
- Aisaka, K.; Hattori, Y.; Kihara, T.; Ishihara, T.; Endo, K.; Hikino, H. Hypotensive action of 3alpha-dihydrocadambine, an indole alkaloid glycoside of uncaria hooks. Planta Med. 1985, 51, 424–427. [Google Scholar] [CrossRef]
- Formagio, A.S.N.; Volobuff, C.R.F.; Kassuya, C.A.L.; Cardoso, C.A.L.; do Carmo Vieira, M.; Pereira, Z.V.; Bagatin, M.C.; de Freitas Gauze, G. Psychotria leiocarpa Extract and Vincosamide Reduce Chemically-Induced Inflammation in Mice and Inhibit the Acetylcholinesterase Activity. Inflammation 2019, 42, 1561–1574. [Google Scholar] [CrossRef]
- Shi, J.S.; Huang, B.; Wu, Q.; Ren, R.X.; Xie, X.L. Effects of rhynchophylline on motor activity of mice and serotonin and dopamine in rat brain. Zhongguo Yao Li Xue Bao 1993, 14, 114–117. [Google Scholar]
- Li, X.M.; Zhang, X.J.; Dong, M.X. Isorhynchophylline Attenuates MPP(+)-Induced Apoptosis through Endoplasmic Reticulum Stress- and Mitochondria-Dependent Pathways in PC12 Cells: Involvement of Antioxidant Activity. Neuromol. Med. 2017, 19, 480–492. [Google Scholar] [CrossRef]
- Zhou, J.; Zhou, S. Antihypertensive and neuroprotective activities of rhynchophylline: The role of rhynchophylline in neurotransmission and ion channel activity. J. Ethnopharmacol. 2010, 132, 15–27. [Google Scholar] [CrossRef]
- Shi, J.S.; Yu, J.X.; Chen, X.P.; Xu, R.X. Pharmacological actions of Uncaria alkaloids, rhynchophylline and isorhynchophylline. Acta Pharmacol. Sin. 2003, 24, 97–101. [Google Scholar] [PubMed]
- Zhou, J.Y.; Zhou, S.W. Isorhynchophylline: A plant alkaloid with therapeutic potential for cardiovascular and central nervous system diseases. Fitoterapia 2012, 83, 617–626. [Google Scholar] [CrossRef] [PubMed]
- Dams-O’Connor, K.; Guetta, G.; Hahn-Ketter, A.E.; Fedor, A. Traumatic brain injury as a risk factor for Alzheimer’s disease: Current knowledge and future directions. Neurodegener. Dis. Manag. 2016, 6, 417–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caruso, A.; Nicoletti, F.; Mango, D.; Saidi, A.; Orlando, R.; Scaccianoce, S. Stress as risk factor for Alzheimer’s disease. Pharmacol. Res. 2018, 132, 130–134. [Google Scholar] [CrossRef]
- Norton, S.; Matthews, F.E.; Barnes, D.E.; Yaffe, K.; Brayne, C. Potential for primary prevention of Alzheimer’s disease: An analysis of population-based data. Lancet Neurol. 2014, 13, 788–794. [Google Scholar] [CrossRef] [Green Version]
- Fu, A.K.; Hung, K.W.; Huang, H.; Gu, S.; Shen, Y.; Cheng, E.Y.; Ip, F.C.; Huang, X.; Fu, W.Y.; Ip, N.Y. Blockade of EphA4 signaling ameliorates hippocampal synaptic dysfunctions in mouse models of Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2014, 111, 9959–9964. [Google Scholar] [CrossRef] [Green Version]
- Li, H.Q.; Ip, S.P.; Yuan, Q.J.; Zheng, G.Q.; Tsim, K.K.W.; Dong, T.T.X.; Lin, G.; Han, Y.; Liu, Y.; Xian, Y.F.; et al. Isorhynchophylline ameliorates cognitive impairment via modulating amyloid pathology, tau hyperphosphorylation and neuroinflammation: Studies in a transgenic mouse model of Alzheimer’s disease. Brain Behav. Immun. 2019, 82, 264–278. [Google Scholar] [CrossRef]
- Li, S.; Fan, T.P.; Jia, W.; Lu, A.; Zhang, W. Network pharmacology in traditional Chinese medicine. Evid. Based Complement. Alternat. Med. 2014, 2014, 138460. [Google Scholar] [CrossRef]
- Hopkins, A.L. Network pharmacology. Nat. Biotechnol. 2007, 25, 1110–1111. [Google Scholar] [CrossRef]
- Hopkins, A.L. Network pharmacology: The next paradigm in drug discovery. Nat. Chem. Biol. 2008, 4, 682–690. [Google Scholar] [CrossRef]
- Chen, X.; Yan, C.C.; Zhang, X.; Zhang, X.; Dai, F.; Yin, J.; Zhang, Y. Drug-target interaction prediction: Databases, web servers and computational models. Brief Bioinform. 2016, 17, 696–712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, R.Z.; Yu, S.J.; Bai, H.; Ning, K. TCM-Mesh: The database and analytical system for network pharmacology analysis for TCM preparations. Sci. Rep. 2017, 7, 2821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, G.; Wang, W.; Wang, X.; Xu, M.; Zhang, L.; Ding, L.; Guo, R.; Shi, Y. Network pharmacology-based strategy to investigate pharmacological mechanisms of Zuojinwan for treatment of gastritis. BMC Complement. Altern. Med. 2018, 18, 292. [Google Scholar] [CrossRef]
- Lyu, M.; Zhou, Z.; Wang, X.; Lv, H.; Wang, M.; Pan, G.; Wang, Y.; Fan, G.; Gao, X.; Feng, Y.; et al. Network Pharmacology-Guided Development of a Novel Integrative Regimen to Prevent Acute Graft-vs.-Host Disease. Front. Pharmacol. 2018, 9, 1440. [Google Scholar] [CrossRef] [Green Version]
- Keiser, M.J.; Setola, V.; Irwin, J.J.; Laggner, C.; Abbas, A.I.; Hufeisen, S.J.; Jensen, N.H.; Kuijer, M.B.; Matos, R.C.; Tran, T.B.; et al. Predicting new molecular targets for known drugs. Nature 2009, 462, 175–181. [Google Scholar] [CrossRef] [Green Version]
- Keiser, M.J.; Roth, B.L.; Armbruster, B.N.; Ernsberger, P.; Irwin, J.J.; Shoichet, B.K. Relating protein pharmacology by ligand chemistry. Nat. Biotechnol. 2007, 25, 197–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drwal, M.N.; Griffith, R. Combination of ligand- and structure-based methods in virtual screening. Drug Discov. Today Technol. 2013, 10, e395–e401. [Google Scholar] [CrossRef] [PubMed]
- Sliwoski, G.; Kothiwale, S.; Meiler, J.; Lowe, E.W., Jr. Computational methods in drug discovery. Pharmacol. Rev. 2014, 66, 334–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, P.; Srivastava, P.; Seth, A.; Tripathi, P.N.; Banerjee, A.G.; Shrivastava, S.K. Comprehensive review of mechanisms of pathogenesis involved in Alzheimer’s disease and potential therapeutic strategies. Prog. Neurobiol. 2019, 174, 53–89. [Google Scholar] [CrossRef]
- Josviak, N.D.; Batistela, M.S.; Souza, R.K.M.; Wegner, N.R.; Bono, G.F.; Sulzbach, C.D.; Simao-Silva, D.P.; Piovezan, M.R.; Souza, R.L.R.; Furtado-Alle, L. Plasma butyrylcholinesterase activity: A possible biomarker for differential diagnosis between Alzheimer’s disease and dementia with Lewy bodies? Int. J. Neurosci. 2017, 127, 1082–1086. [Google Scholar] [CrossRef]
- Macdonald, I.R.; Maxwell, S.P.; Reid, G.A.; Cash, M.K.; DeBay, D.R.; Darvesh, S. Quantification of Butyrylcholinesterase Activity as a Sensitive and Specific Biomarker of Alzheimer’s Disease. J. Alzheimer’s Dis. 2017, 58, 491–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kushida, H.; Fukutake, M.; Tabuchi, M.; Katsuhara, T.; Nishimura, H.; Ikarashi, Y.; Kanitani, M.; Kase, Y. Simultaneous quantitative analyses of indole and oxindole alkaloids of Uncaria Hook in rat plasma and brain after oral administration of the traditional Japanese medicine Yokukansan using high-performance liquid chromatography with tandem mass spectrometry. Biomed. Chromatogr. 2013, 27, 1647–1656. [Google Scholar] [CrossRef] [PubMed]
- Han, A.; Lin, G.; Cai, J.; Wu, Q.; Geng, P.; Ma, J.; Wang, X.; Lin, C. Pharmacokinetic study on hirsutine and hirsuteine in rats using UPLC–MS/MS. Acta Chromatogr. 2019, 31, 99–104. [Google Scholar] [CrossRef]
- Wang, J. The Metabolic Study of Indole Alkaloids in Uncaria Rhynchophylla In Vivo; China Pharmaceutical University: Nanjing, China, 2017. [Google Scholar]
- Lagorce, D.; Bouslama, L.; Becot, J.; Miteva, M.A.; Villoutreix, B.O. FAF-Drugs4: Free ADME-tox filtering computations for chemical biology and early stages drug discovery. Bioinformatics 2017, 33, 3658–3660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, Z.J.; Dong, J.; Che, Y.J.; Zhu, M.F.; Wen, M.; Wang, N.N.; Wang, S.; Lu, A.P.; Cao, D.S. TargetNet: A web service for predicting potential drug-target interaction profiling via multi-target SAR models. J. Comput. Aided Mol. Des. 2016, 30, 413–424. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef] [Green Version]
- Gfeller, D.; Michielin, O.; Zoete, V. Shaping the interaction landscape of bioactive molecules. Bioinformatics 2013, 29, 3073–3079. [Google Scholar] [CrossRef]
- Wang, X.; Shen, Y.; Wang, S.; Li, S.; Zhang, W.; Liu, X.; Lai, L.; Pei, J.; Li, H. PharmMapper 2017 update: A web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 2017, 45, W356–W360. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Ouyang, S.; Yu, B.; Liu, Y.; Huang, K.; Gong, J.; Zheng, S.; Li, Z.; Li, H.; Jiang, H. PharmMapper server: A web server for potential drug target identification using pharmacophore mapping approach. Nucleic Acids Res. 2010, 38, W609–W614. [Google Scholar] [CrossRef] [Green Version]
- Bardou, P.; Mariette, J.; Escudie, F.; Djemiel, C.; Klopp, C. jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Cheng, D.; Shrivastava, S.; Tzur, D.; Gautam, B.; Hassanali, M. DrugBank: A knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008, 36, D901–D906. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ji, Z.L.; Chen, Y.Z. TTD: Therapeutic Target Database. Nucleic Acids Res. 2002, 30, 412–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef] [Green Version]
- Kamburov, A.; Stelzl, U.; Lehrach, H.; Herwig, R. The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res. 2013, 41, D793–D800. [Google Scholar] [CrossRef]
- Slenter, D.N.; Kutmon, M.; Hanspers, K.; Riutta, A.; Windsor, J.; Nunes, N.; Melius, J.; Cirillo, E.; Coort, S.L.; Digles, D.; et al. WikiPathways: A multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 2018, 46, D661–D667. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res. 2019, 47, D1102–D1109. [Google Scholar] [CrossRef] [Green Version]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellman, G.L.; Courtney, K.D.; Andres, V., Jr.; Feather-Stone, R.M. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 1961, 7, 88–95. [Google Scholar] [CrossRef]










| # | Gene Symbol | Degree | Subgraph | Eigenvector | Information | LAC | Betweenness | Closeness | Network |
|---|---|---|---|---|---|---|---|---|---|
| 1 | MAOA | 16 | 1850.135 | 0.356 | 5.360 | 6.375 | 81.651 | 0.759 | 13.078 |
| 2 | ACHE | 14 | 1413.283 | 0.311 | 5.162 | 5.143 | 63.508 | 0.710 | 9.577 |
| 3 | BCHE | 13 | 1311.436 | 0.300 | 5.048 | 4.923 | 38.836 | 0.688 | 8.028 |
| 4 | DRD2 | 12 | 1230.716 | 0.290 | 4.924 | 6.333 | 35.935 | 0.667 | 9.991 |
| 5 | HTR1A | 11 | 1166.267 | 0.282 | 4.786 | 6.545 | 20.340 | 0.629 | 9.071 |
| 6 | HTR2A | 10 | 623.717 | 0.203 | 4.632 | 4.800 | 24.033 | 0.595 | 7.526 |
| 7 | ADRA2B | 9 | 868.194 | 0.242 | 4.461 | 6.444 | 7.798 | 0.595 | 8.000 |
| 8 | ADRA2C | 9 | 868.194 | 0.242 | 4.461 | 6.444 | 7.798 | 0.595 | 8.000 |
| 9 | ADRA2A | 9 | 868.194 | 0.242 | 4.461 | 6.444 | 7.798 | 0.595 | 8.000 |
| 10 | CYP3A4 | 9 | 688.133 | 0.216 | 4.461 | 4.889 | 10.826 | 0.629 | 6.405 |
| 11 | ALB | 8 | 406.076 | 0.165 | 4.268 | 2.750 | 47.899 | 0.611 | 3.429 |
| 12 | CYP2D6 | 8 | 606.406 | 0.203 | 4.268 | 4.500 | 9.607 | 0.611 | 5.238 |
| 13 | ADRA1D | 7 | 428.160 | 0.169 | 4.049 | 4.857 | 1.567 | 0.537 | 6.000 |
| 14 | ADRA1B | 7 | 428.160 | 0.169 | 4.049 | 4.857 | 1.567 | 0.537 | 6.000 |
| 15 | ADRA1A | 7 | 428.160 | 0.169 | 4.049 | 4.857 | 1.567 | 0.537 | 6.000 |
| 16 | CYP2C19 | 7 | 271.666 | 0.133 | 4.049 | 3.429 | 39.552 | 0.564 | 4.333 |
| 17 | CHRM2 | 6 | 348.284 | 0.151 | 3.798 | 5.000 | 0.000 | 0.458 | 6.000 |
| 18 | CHRM4 | 6 | 348.284 | 0.151 | 3.798 | 5.000 | 0.000 | 0.458 | 6.000 |
| 19 | CHRM5 | 5 | 178.487 | 0.107 | 3.508 | 4.000 | 0.000 | 0.512 | 5.000 |
| 20 | CYP1A2 | 5 | 193.747 | 0.112 | 3.508 | 3.200 | 1.269 | 0.537 | 4.000 |
| 21 | DRD1 | 4 | 252.070 | 0.131 | 3.170 | 2.500 | 0.167 | 0.524 | 3.333 |
| 22 | PPARG | 2 | 7.872 | 0.019 | 2.287 | 0.000 | 2.700 | 0.400 | 0.000 |
| 23 | PTGS1 | 2 | 6.536 | 0.016 | 2.287 | 0.000 | 1.583 | 0.379 | 0.000 |
| Ligand | Total Scores |
|---|---|
| 3F9_A | 5.524 |
| 3F9_B | 4.6765 |
| Corynoxine B | 5.7701 |
| Corynoxeine (C) | 5.4715 |
| Corynoxine | 5.58 |
| 3-dihydrocadambine (DHC) | 5.0741 |
| Geissoschiziner (GSM) | 5.886 |
| Hirsuteine (HTE) | 6.7146 |
| Hirsutine (HTI) | 6.4769 |
| Isocorynoxeine (IC) | 6.196 |
| Isorhynchophylline (IR) | 6.1038 |
| isomitraphylline | 4.3782 |
| Rhynchophylline (R) | 4.8015 |
| Uncarine B | 4.8631 |
| Vincosamide (VCA) | 5.9499 |
| Galantamine hydrobromide | 4.8609 |
| Drofenine hydrochloride | 9.7639 |
| Conc. (μM) | Inhibition Rate (%) | ||||
|---|---|---|---|---|---|
| Hirsutine (HTI) | Hirsuteine (HTE) | Geissoschiziner (GSM) | Corynoxeine | Isocorynoxeine | |
| 50 | 50.71 ± 1.51 | 21.23 ± 0.74 | 0.07 ± 0.99 | - | - |
| 100 | 65.83 ± 2.49 | 36.03 ± 0.67 | 7.34 ± 0.66 | - | - |
| 200 | 71.69 ± 0.18 | 54.44 ± 0.92 | 22.24 ± 1.32 | (−1.86) ± 1.65 | (−1.86) ± 0.55 |
| 300 | 73.28 ± 0.67 | 60.51 ± 0.74 | n.a. | 1.76 ± 0.98 | 3.09 ± 1.58 |
| 400 | 73.60 ± 0.67 | 64.13 ± 0.18 | n.a. | 16.82 ± 3.39 | 15.17 ± 3.50 |
| 500 | 73.17 ± 1.15 | 65.19 ± 0.96 | 39.83 ± 1.52 | 19.43 ± 3.21 | 21.77 ± 0.90 |
| IC50 | 67.97 | 202.3 | n.a. | n.a. | n.a. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, W.; Zhang, Z.; Li, F.; Deng, Y.; Lei, M.; Long, H.; Hou, J.; Wu, W. A Network-Based Approach to Explore the Mechanisms of Uncaria Alkaloids in Treating Hypertension and Alleviating Alzheimer’s Disease. Int. J. Mol. Sci. 2020, 21, 1766. https://doi.org/10.3390/ijms21051766
Wu W, Zhang Z, Li F, Deng Y, Lei M, Long H, Hou J, Wu W. A Network-Based Approach to Explore the Mechanisms of Uncaria Alkaloids in Treating Hypertension and Alleviating Alzheimer’s Disease. International Journal of Molecular Sciences. 2020; 21(5):1766. https://doi.org/10.3390/ijms21051766
Chicago/Turabian StyleWu, Wenyong, Zijia Zhang, Feifei Li, Yanping Deng, Min Lei, Huali Long, Jinjun Hou, and Wanying Wu. 2020. "A Network-Based Approach to Explore the Mechanisms of Uncaria Alkaloids in Treating Hypertension and Alleviating Alzheimer’s Disease" International Journal of Molecular Sciences 21, no. 5: 1766. https://doi.org/10.3390/ijms21051766