Illumina® Sequencing Reveals Candidate Genes of Carotenoid Metabolism in Three Pummelo Cultivars (Citrus Maxima) with Different Pulp Color
Abstract
:1. Introduction
2. Results
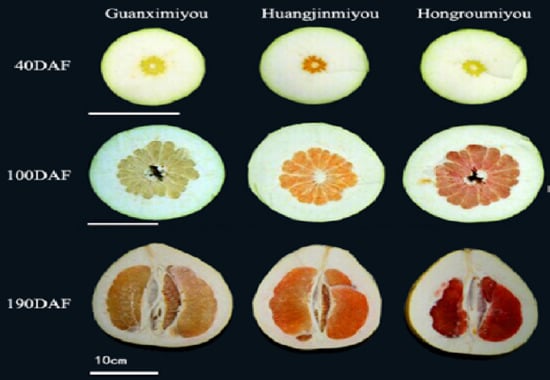
2.1. Changes in β-Carotene and Lycopene Contents During Fruit Development of Three Different Pummelo Cultivars
2.2. Transcriptome Sequence and De Novo Assembly
2.3. Functional Annotation and Classification of Unigenes
2.4. Analysis of DEG During Fruit Development of Three Different Pummelo Cultivars
2.5. Candidate Genes Involved in Carotenoid Biosynthesis
2.6. Real-Time Quantitative PCR (RT-qPCR) Analysis of Selected DEGs During Fruit Development of Three Pummelo Cultivars
3. Discussion
4. Material and Methods
4.1. Plant Materials
4.2. β-Carotene and Lycopene Extractions and Analysis by HPLC
4.3. RNA Extraction, cDNA Library Construction and Sequencing
4.4. Reads Assembly and Functional Annotation
4.5. Identification of Genes Related to the Carotenoid Biosynthesis Pathways and Transcription Factor
4.6. Correlation Analysis of Structural Genes and Transcription Factors
4.7. RT-qPCR analysis
4.8. Experimental Design and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nisar, N.; Li, L.; Lu, S.; Khin, N.C.; Pogson, B.J. Carotenoid metabolism in plants. Mol. Plant 2015, 8, 68–82. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Ikoma, Y.; Matsumoto, H.; Sugiura, M.; Hyodo, H.; Yano, M. Accumulation of carotenoids and expression of carotenoid biosynthetic genes during maturation in citrus fruit. Plant Physiol. 2004, 134, 824–837. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Luo, T.; Liu, C.Y.; Wang, Y.; Yang, H.B.; Yang, W.; Zheng, L.; Xiao, X.; Zhang, M.F.; Xu, R.W.; et al. An R2R3-MYB transcription factor represses the transformation of alpha- and beta-branch carotenoids by negatively regulating expression of CrBCH2 and CrNCED5 in flavedo of Citrus reticulate. New Phytol. 2017, 216, 178–192. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.W.; Zhang, Y.; Zhu, K.J.; Yang, W.; Ye, J.L.; Chai, L.J.; Xu, Q.; Deng, X.X. The citrus transcription factor CsMADS6 modulates carotenoid metabolism by directly regulating carotenogenic genes. Plant Physiol. 2018. [Google Scholar] [CrossRef]
- Ikoma, Y.; Matsumoto, H.; Kato, M. Diversity in the carotenoid profiles and the expression of genes related to carotenoid accumulation among citrus genotypes. Breed. Sci. 2016, 66, 139–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigo, M.J.; Alquézar, B.; Alós, E.; Lado, J.; Zacarías, L. Biochemical bases and molecular regulation of pigmentation in the peel of Citrus fruit. Sci. Hortic. 2013, 163, 46–62. [Google Scholar] [CrossRef]
- Yu, K.Q.; Xu, Q.; Da, X.L.; Guo, F.; Ding, Y.D.; Deng, X.X. Transcriptome changes during fruit development and ripening of sweet orange (Citrus sinensis). Bmc Genom. 2012, 13, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Xu, Z.; Zhang, Y.; Chai, L.; Yi, H.; Deng, X.X. An integrative analysis of the transcriptome and proteome of the pulp of a spontaneous late-ripening sweet orange mutant and its wild type improves our understanding of fruit ripening in citrus. J. Exp. Bot. 2014, 65, 1651–1671. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.J.; Wang, X.J.; Wu, J.X.; Chen, S.Y.; Chen, H.; Chai, L.J.; Yi, H.L. Comparative transcriptome analyses between a spontaneous late ripening sweet orange mutant and its wild type suggest the functions of ABA, sucrose and JA during citrus fruit ripening. PLoS ONE 2014, 9, 116056. [Google Scholar] [CrossRef]
- Guo, F.; Yu, H.; Xu, Q.; Deng, X. Transcriptomic analysis of differentially expressed genes in an orange-pericarp mutant and wild type in pummelo (Citrus grandis). Bmc Plant Biol. 2015, 15, 44. [Google Scholar] [CrossRef]
- Liu, C.H.; Yan, F.H.; Gao, H.J.; He, M.; Wang, Z.; Cheng, Y.J.; Deng, X.X.; Xu, J. Features of citrus terpenoid production as revealed by carotenoid, limonoid and aroma profiles of two pummelos (Citrus maxima) with different flesh color. J. Sci. Food Agric. 2015, 95, 111–119. [Google Scholar] [CrossRef]
- Lu, X.K.; Lin, Q.H.; Lu, X.M.; Zhang, S.M.; Li, C.S.; Ye, X.F. Comparison of carotenoid compositions and contents n different sweet pomelos. Fujianj Agric. Sci. 2012, 27, 723–727. [Google Scholar]
- Lu, X.K.; Lin, Q.H.; Lin, Y.J.; Zhang, J.T.; Zhang, S.M.; Li, C.S. ‘Huangjinmiyou’, a new orange-yellow fleshed pomelo cultivar. J. Fruit Sci. 2013, 30, 900–902. [Google Scholar]
- BLASTX and BLASTN searching engines. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi.
- Ampomah-Dwamena, C.; McGhie, T.; Wibisono, R.; Montefiori, M.; Hellens, R.P.; Allan, A.C. The kiwifruit lycopene beta-cyclase plays a significant role in carotenoid accumulation in fruit. J. Exp. Bot. 2009, 60, 3765–3779. [Google Scholar] [CrossRef] [Green Version]
- Lv, P.; Li, N.; Gu, H.H.; Zhao, W.E. Changes in carotenoid profiles and in the expression pattern of the genes in carotenoid metabolisms during fruit development and ripening in four watermelon cultivar. Food Chem. 2015, 174, 52–59. [Google Scholar] [CrossRef]
- Mizuno, K.; Tokiwano, T.; Yoshizawa, Y. Gene expression analysis of enzymes of the carotenoid biosynthesis pathway involved in β-cryptoxanthin accumulation in wild raspberry, Rubus palmatus. Biochem. Biophys. Commun. 2017, 484, 845–849. [Google Scholar] [CrossRef] [PubMed]
- Leng, X.P.; Wang, P.P.; Wang, C.; Zhu, X.D.; Li, X.P.; Li, H.Y.; Mu, Q.M.; Liu, Z.J.; Fang, J.G. Genome-wide identification and characterization of genes involved in carotenoid metabolic in three stages of grapevine fruit development. Sci. Rep. 2017, 7, 4216. [Google Scholar] [CrossRef] [Green Version]
- Richaud, D.; Stange, C.; Gadaleta, A.; Colasuonno, P.; Parada, R.; Schwember, A.R. Identification of Lycopene epsilon cyclase (LCYE) gene mutants to potentially increase β-carotene content in durum wheat (Triticum turgidum L.ssp. Durum) through TILLING. PLoS ONE 2018, 13, e0208948. [Google Scholar]
- Colasuonno, P.; Lozito, M.L.; Marcotuli, I.; Nigro, D.; Giancaspro, A.; Mangini, G.; De Vita, P.; Mastrangelo, A.M.; Pecchioni, N.; Houston, K.; et al. The carotenoid biosynthetic and catabolic genes in wheat and their association with yellow pigments. Bmc Genom. 2017, 18, 122. [Google Scholar] [CrossRef]
- Colasuonno, P.; Incerti, O.; Lozito, M.L.; Simeone, R.; Gadaleta, A.; Blanco, A. DHPLC technology for high-throughput detection of mutations in a durum wheat TILLING population. Bmc Genet. 2016, 17, 43. [Google Scholar] [CrossRef] [PubMed]
- Charles, A.D.; Driedonks, N.; Lewis, D.; Shumskaya, M.; Chen, X.Y.; Wurtzel, E.T.; Espley, R.V.; Allan, A.C. The Phytoene synthase gene family of apple (Malus x domestica) and its role in controlling fruit carotenoid content. Bmc Plant Biol. 2015, 15, 185. [Google Scholar]
- Clotault, J.; Peltier, D.; Berruyer, R.; Thomas, M.; Briard, M.; Geoffriau, E. Expression of carotenoid biosynthesis genes during carrot root development. J. Exp. Bot. 2008, 59, 3563–3573. [Google Scholar] [CrossRef] [Green Version]
- Fraser, P.D.; Eugenia, M.A.A.; Halket, J.M.; Truesdale, M.R.; Yu, D.M.; Gerrish, C.; Bramley, P.M. Manipulation of phytoene levels in tomato fruit: Effects on isoprenoids; plastids; and intermediary metabolism. Plant Cell 2007, 19, 3194–3211. [Google Scholar] [CrossRef]
- Rodrigo, M.J.; Marcos, J.F.; Zacarias, L. Biochemical and molecular analysis of carotenoid biosynthesis in flavedo of orange (Citrus sinensis L.) during fruit development and maturation. J. Agric. Food Chem. 2004, 52, 6724–6731. [Google Scholar] [CrossRef]
- Wei, X.; Chen, C.X.; Yu, Q.B.; Gady, A.; Yu, Y.; Liang, G.L.; Gmitter, F.G. Comparison of carotenoid accumulation and biosynthesis gene expression between Valencia and Rohde Red Valencia sweet oranges. Plant Sci. 2014, 227, 28–36. [Google Scholar] [CrossRef]
- Guo, F.; Zhou, W.; Zhang, J.; Xu, Q.; Deng, X.X. Effect of the citrus lycopene β-cyclase transgene on carotenoid metabolism in transgenic tomato fruits. PLoS ONE 2012, 7, e32221. [Google Scholar] [CrossRef] [PubMed]
- Mendes, A.F.S.; Chen, C.X.; Gmitter, F.G.; Moore, G.A.; Costa, M.G.C. Expression and phylogenetic analysis of two new lycopene β-cyclases from Citrus paradisi. Physiol. Plant 2011, 141, 1–10. [Google Scholar] [CrossRef]
- Chen, C.; Costa, M.G.C.; Yu, Q.; Moore, G.C.; Gmitter, F.G. Identification of novel members in sweet orange carotenoid biosynthesis gene families. Tree Genet. Genome 2010, 6, 905–914. [Google Scholar] [CrossRef]
- Kang, C.; Zhai, H.; Xue, L.Y.; Zhao, N.; He, S.Z.; Liu, Q.C. A lycopene β-cyclase gene; IbLCYB2; enhances carotenoid contents and abiotic stress tolerance in transgenic sweetpotato. Plant Sci. 2018, 272, 243–254. [Google Scholar] [CrossRef]
- Devitt, L.C.; Fanning, K.; Dietzgen, R.G.; Holton, T.A. Isolation and functional characterization of a lycopene β-cyclase gene that controls fruit colour of papaya (Carica papaya L.). J. Exp. Bot. 2010, 61, 33. [Google Scholar] [CrossRef] [PubMed]
- Auldridge, M.E.; McCarty, D.R.; Klee, H.J. Plant carotenoid cleavage oxygenases and their apocarotenoid products. Curr. Opin. Plant Biol. 2006, 9, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Bouvier, F.; Suire, C.; Mutterer, J.; Camara, B. Oxidative remodeling of chromoplast carotenoids: Identification of the carotenoid dioxygenase CsCCD and CsZCD genes involved in Crocus secondary metabolite biogenesis. Plant Cell 2003, 15, 47–62. [Google Scholar] [CrossRef]
- Yoshioka, S.; Aida, R.; Yamamizo, C.; Shibata, M.; Ohmiya, A. The carotenoid cleavage dioxygenase 4 (CmCCD4a) gene family encodes a key regulator of petal color mutation in chrysanthemum. Euphytica 2012, 184, 377–387. [Google Scholar] [CrossRef]
- Ureshino, K.; Nakayaa, M.; Miyajima, I. Contribution made by the carotenoid cleavage dioxygenase 4 gene to yellow colour fade in azalea petals. Euphytica 2016, 207, 401–417. [Google Scholar] [CrossRef]
- Song, C.; Zhang, L.; Zhang, C.; Tian, Y.; Cong, P. Differential expression analysis of the fruits of yellow-fleshed apple and exploits genes involved in carotenoid pigmentation. Acta Hortic. Sin. 2017. [Google Scholar] [CrossRef]
- Bai, S.; Tuan, P.A.; Tatsuki, M.; Yaegaki, H.; Ohmiya, A.; Yamamizo, C.; Moriguchi, T. Knockdown of carotenoid cleavage dioxygenase 4 (CCD4) via virus-induced gene silencing confers yellow coloration in peach fruit: Evaluation of gene function related to fruit traits. Plant Mol. Biol. Rep. 2016, 34, 257–264. [Google Scholar] [CrossRef]
- Bruno, M.; Beyer, P.; Al-Babili, S. The potato carotenoid cleavage dioxygenase 4 catalyzes a single cleavage of biononering-containing carotenes and non-epoxidated xanthophylls. Arch. Biochem. Biophys. 2015, 572, 126–133. [Google Scholar] [CrossRef]
- González-Verdejo, C.; Obrero, A.; Román, B.; Gómez, P. Expression profile of carotenoid cleavage dioxygenase genes in summer squash (Cucurbita pepo L.). Plant Foods Hum. Nutr. 2015, 70, 200–206. [Google Scholar]
- Schwartz, S.H.; Qin, X.; Zeevaart, J.A. Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol. 2003, 131, 1591–1601. [Google Scholar] [CrossRef]
- Barickman, T.C.; Kopsell, D.A.; Sams, C.E. Abscisic acid increases carotenoid and chlorophyll concentrations in leaves and fruit of two tomato genotypes. J. Am. Soc. Hortic. Sci. 2014, 139, 261–266. [Google Scholar] [CrossRef]
- Cantin, C.M.; Fidelibus, M.W.; Crisostoc, C.H. Application of abscisic acid (ABA) at veraison advanced red color development and maintained postharvest quality of ‘Crimson Seedless’ grapes. Postharvest Biol. Technol. 2007, 46, 237–241. [Google Scholar] [CrossRef] [Green Version]
- Colasuonno, P.; Marcotuli, I.; Lozito, M.L.; Simeone, R.; Blanco, A.; Gadaleta, A. Characterization of aldehyde oxidase (AO) genes involved in the accumulation of carotenoid pigments in wheat grain. Bmc Genet. 2017, 8, 863. [Google Scholar] [CrossRef]
- Chernys, J.T.; Zeevaart, J.A. Characterization of the 9-cis-epoxycarotenoid dioxygenase gene family and the regulation of abscisic acid biosynthesis in avocado. Plant Physiol. 2000, 124, 343–353. [Google Scholar] [CrossRef]
- Meier, S.; Tzfadia, O.; Vallabhaneni, R.; Gehring, C.; Wurtzel, E.T. A transcriptional analysis of carotenoid; chlorophyll and plastidial isoprenoid biosynthesis genes during development and osmotic stress responses in Arabidopsis thaliana. Bmc Syst. Biol. 2011, 5, 77. [Google Scholar] [CrossRef]
- Sun, L.; Yuan, B.; Zhang, M.; Wang, L.; Cui, M.; Wang, Q.; Leng, P. Fruit-specific RNAi-mediated suppression of SlNCED1 increases both lycopene and beta-carotene contents in tomato fruit. J. Exp. Bot. 2012, 63, 3097–3108. [Google Scholar] [CrossRef]
- Fang, J.; Chai, C.; Qian, Q.; Li, C.; Tang, J.; Sun, L.; Huang, Z.; Guo, X.; Sun, C.; Liu, M.; et al. Mutations of genes in synthesis of the carotenoid precursors of ABA lead to preharvest sprouting and photooxidation in rice. Plant J. 2008, 54, 177–189. [Google Scholar] [CrossRef]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef]
- Ji, K.; Kai, W.; Zhao, B.; Sun, Y.; Yuan, B.; Dai, S.; Li, Q.; Chen, P.; Wang, Y.; Pei, Y.; et al. SlNCED1 and SlCYP707A2: Key genes involved in ABA metabolism during tomato fruit ripening. J. Exp. Bot. 2014, 65, 5243–5255. [Google Scholar] [CrossRef]
- Li, D.D.; Li, L.; Luo, Z.S.; Mou, W.S.; Mao, L.C.; Ying, T.J. Comparative transcriptome analysis reveals the influence of abscisic acid on the metabolism of pigments; ascorbic acid and folic acid during strawberry fruit ripening. PLoS ONE 2015, 10, e0130037. [Google Scholar] [CrossRef]
- Welsch, R.; Beyer, P.; Hugueney, P.; Kleinig, H.; von Lintig, J. Regulation and activation of phytoene synthase, a key enzyme in carotenoid biosynthesis, during photomorphogenesis. Planta 2000, 211, 846–854. [Google Scholar] [CrossRef]
- Toledo-Ortiz, G.; Huq, E.; Rodriguez-Concepcion, M. Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 2010, 107, 11626–11631. [Google Scholar] [CrossRef] [Green Version]
- Grassi, S.; Piro, G.; Lee, J.M.; Zheng, Y.; Fei, Z.J.; Dalessandro, G.; Giovannoni, J.J.; Lenucci, M.S. Comparative genomics reveals candidate carotenoid pathway regulators of ripening watermelon fruit. Bmc Genom. 2013, 14, 781. [Google Scholar] [CrossRef]
- Li, R.; Zhai, H.; Kang, C.; Liu, D.; He, S.Z.; Liu, Q.C. De Novo transcriptome sequencing of the orange-fleshed sweet potato and analysis of differentially expressed genes related to carotenoid biosynthesis. Int. J. Genom. 2015, 2015, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.M.; Joung, J.G.; McQuinn, R.; Chung, M.Y.; Fei, Z.; Tieman, D.; Klee, H.; Giovannoni, J.J. Combined transcriptome; genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. Plant J. 2012, 70, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.K.; Chen, G.P.; Zhou, S.; Tu, Y.; Wang, Y.; Dong, T.T.; Hu, Z.L. A new tomato NAC (NAM/ATAF1/2/CUC2) transcription factor; SlNAC4; functions as a positive regulator of fruit ripening and carotenoid accumulation. Plant Cell Physiol. 2014, 55, 119–135. [Google Scholar] [CrossRef] [PubMed]
- Llorente, B.D.; Andrea, L.; Ruizsola, M.A.; Botterweg, E.; Pulido, P. Tomato fruit carotenoid biosynthesis is adjusted to actual ripening progression by a light-dependent mechanism. Plant J. 2016, 85, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Endo, T.; Fujii, H.; Sugiyama, A.; Nakano, M.; Nakajima, N.; Ikoma, Y.; Omura, M.; Shimada, T. Overexpression of a citrus basic helix-loop-helix transcription factor (CubHLH1), which is homologous to Arabidopsis activation-tagged bri1 suppressor 1 interacting factor genes; modulates carotenoid metabolism in transgenic tomato. Plant Sci. 2016, 243, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.G.; Chen, G.P.; Guo, X.H.; Yin, W.C.; Yu, X.H.; Hu, J.T.; Hu, Z.L. Overexpression of SlPRE2, an atypical bHLH transcription factor; affects plant morphology and fruit pigment accumulation in tomato. Sci. Rep. 2017, 7, 5786. [Google Scholar] [CrossRef]
- Banerjee, A.; Roychoudhury, A. WRKY proteins: Signaling and regulation of expression during abiotic stress responses. Sci. World J. 2015, 807560. [Google Scholar] [CrossRef]
- Han, Y.J.; Wu, M.; Cao, L.Y.; Yuan, W.J.; Dong, M.; Wang, X.H.; Chen, W.C.; Shang, F.D. Characterization of Of WRKY3; a transcription factor that positively regulates the carotenoid cleavage dioxygenase gene Of CCD4 in Osmanthus fragrans. Plant Mol. Biol. 2016, 91, 485–496. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.N.; Feng, H.L.; Meng, X.; Li, D.; Yang, D.Y.; Wu, C.G.; Meng, Q.W. Overexpression of tomato SlNAC1 transcription factor alters fruit pigmentation and softening. Bmc Plant Biol. 2014, 14, 351. [Google Scholar] [CrossRef]
- Fu, C.C.; Han, Y.C.; Fan, Z.Q.; Chen, J.Y.; Chen, W.X.; Lu, W.J.; Kuang, J.F. The papaya transcription factor cpnac1 modulates carotenoid biosynthesis through activating phytoene desaturase genes CpPDS2/4 during fruit ripening. J. Agric. Food Chem. 2016, 64, 5454. [Google Scholar] [CrossRef] [PubMed]
- Kou, X.H.; Zhao, Y.N.; Wu, C.; Jiang, B.L.; Zhang, Z.; Rathbun, J.R.; He, Y.L.; Xue, Z.H. SNAC4 and SNAC9 transcription factors show contrasting effects on tomato carotenoids biosynthesis and softening. Postharvest Biol. Tech. 2018, 144, 9–19. [Google Scholar] [CrossRef]
- Giovannoni, J.J. Genetic regulation of fruit development and ripening. Plant Cell 2004, 16, S170–S180. [Google Scholar] [CrossRef] [PubMed]
- Welsch, R.; Maass, D.; Voegel, T.; Dellapenna, D.; Beyer, P. Transcription factor RAP2.2 and its interacting partner SINAT2: Stable elements in the carotenogenesis of Arabidopsis leaves. Plant Physiol. 2007, 45, 1073–1085. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.C.; Han, Y.C.; Kuang, J.F.; Chen, J.Y.; Lu, W.J. Papaya CpEIN3a and CpNAC2 cooperatively regulate carotenoid biosynthesis related genes CpPDS2/4, CpLCY-e, and CpCHY-b during fruit ripening. Plant Cell Physiol. 2017, 58, 2155–2165. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Shimada, T.; Sugiyama, A.; Nishikawa, F.; Endo, T.; Nakano, M.; Ikoma, Y.; Shimizu, T.; Omura, M. Profiling ethylene-responsive genes in mature mandarin fruit using a citrus 22K oligoarray. Sci. Hortic. 2007, 173, 340–348. [Google Scholar] [CrossRef]
- Allan, A.C.; Hellens, R.P.; Laing, W.A. MYB transcription factors that colour our fruit. Trends Plant Sci. 2008, 13, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Deluc, L.; Bogs, J.; Walker, A.R.; Ferrier, T.; Decendit, A.; Merillon, J.M.; Robinson, S.P.; Barrieu, F. The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant Physiol. 2008, 147, 2041–2053. [Google Scholar] [CrossRef] [PubMed]
- Martins, T.R.; Jiang, P.; Rausher, M.D. How petals change their spots: Cis-regulatory re-wiring in Clarkia (Onagraceae). New Phytol. 2016, 216, 510. [Google Scholar] [CrossRef]
- Sagawa, J.M.; Stanley, L.E.; LaFountain, A.M.; Frank, H.A.; Liu, C.; Yuan, Y.W. An R2R3-MYB transcription factor regulates carotenoid pigmentation in Mimulus lewisii flowers. New Phytol. 2016, 209, 1049–1057. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Tao, N.G.; Liu, Q.; Deng, X.X. Presence of diverse rations of lycopene β–carotene in five pink or red-fleshed citrus cultivars. Sci. Hortic. 2006, 108, 181–184. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C. RSEM: Accurate transcript quantification from RNASeq data with or without a reference genome. Bmc Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- NCBI database. Available online: https://www.ncbi.nlm.nih.gov/sra.
- PlantTFDB 3.0 software. Available online: http://planttfdb.cbi.pku.edu.cn/prediction.php.








| Samples | Read number | Base number | GC content | % (≥Q30) |
|---|---|---|---|---|
| GXMY1 (40 DAF) | 22,744,151 | 4,581,699,084 | 46.49% | 85.92% |
| GXMY2 (100 DAF) | 22,575,178 | 4,548,053,121 | 46.10% | 85.79% |
| GXMY3 (190 DAF) | 22,780,099 | 4,589,605,411 | 45.77% | 85.82% |
| HRMY1 (40 DAF) | 22,437,953 | 4,515,689,216 | 47.34% | 85.73% |
| HRMY2 (100 DAF) | 22,964,175 | 4,624,970,836 | 45.06% | 86.05% |
| HRMY3 (190 DAF) | 22,607,952 | 4,557,243,046 | 44.76% | 85.88% |
| HJMY1 (40 DAF) | 22,064,221 | 4,450,273,681 | 46.50% | 85.68% |
| HJMY2 (100 DAF) | 19,694,950 | 3,977,639,966 | 45.25% | 85.02% |
| HJMY3 (190 DAF) | 23,914,670 | 4,816,251,310 | 45.48% | 86.02% |
| Total | 201,783,349 | 40,661,425,671 | 45.86% | 85.76% |
| After assembly | ||||
| Total clean reads | 201,783,349 | |||
| Total clean nucleotides (nt) | 40,661,425,671 | |||
| Total number of contigs | 17,658,287 | |||
| Total length of contigs (nt) | 656,824,421 | |||
| Mean length of contigs (nt) | 37 | |||
| N50 of contigs | 37 | |||
| Total number of unigenes | 54,051 | |||
| Total length of unigenes (nt) | 46,385,997 | |||
| Mean length of unigenes (nt) | 858 | |||
| N50 of unigenes | 1,585 | |||
| Fruit development at different stages (days) | GXMY | HRMY | HJMY | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ALL DEGs | Up-regulated | Down-regulated | ALL DEGs | Up-regulated | Down-regulated | ALL DEGs | Up-regulated | Down-regulated | |
| 40 DAF/100 DAF | 3240 | 1068 | 2172 | 1887 | 371 | 1516 | 2080 | 421 | 1,659 |
| 40 DAF/190 DAF | 3882 | 1408 | 2474 | 2569 | 753 | 1816 | 2316 | 639 | 1,677 |
| 100 DAF/190 DAF | 2562 | 1300 | 1262 | 4988 | 1875 | 3113 | 3371 | 1796 | 1,575 |
| 40 DAF/100 DAF/190 DAF | 616 | 258 | 256 | ||||||
| Gene Name | Unigene ID | Gene Length | GXMY | HRMY | HJMY | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 40 DAF | 100 DAF | 190 DAF | 40 DAF | 100 DAF | 190 DAF | 40 DAF | 100 DAF | 190 DAF | |||
| PSY2 | c27977.graph_c0 | 1373 | 3.45 | 0.41 | 0.04 | 0.04 | 0.32 | 0 | 0 | 0.28 | 0.04 |
| LYCB2 | c35136.graph_c0 | 2037 | 5.57 | 15.15 | 19.49 | 2.3 | 15.07 | 25.57 | 0.58 | 15.4 | 19.32 |
| LYCE | c29745.graph_c0 | 2018 | 6.22 | 4.01 | 1.64 | 0.71 | 6.95 | 4.68 | 0.63 | 7.78 | 3.09 |
| CCD1 | c29184.graph_c0 | 903 | 6.04 | 3.81 | 0.65 | 1.4 | 5.87 | 0.47 | 1.26 | 5.31 | 0 |
| CCD4 | c20564.graph_c0 | 3057 | 60.55 | 20.59 | 16.36 | 96.28 | 18.75 | 16.19 | 118.99 | 17.24 | 12.32 |
| NCED1 | c17328.graph_c1 | 1536 | 8.23 | 0.45 | 5.20 | 12.65 | 1.09 | 10.87 | 2.23 | 0.63 | 8.46 |
| NCED2 | c31914.graph_c0 | 2425 | 17.56 | 107.12 | 99.71 | 10.13 | 105.11 | 84.13 | 4.57 | 97.99 | 59.51 |
| NCED3 | c11942.graph_c0 | 1101 | 11.74 | 0.46 | 7.28 | 14.57 | 0.51 | 10.62 | 3.26 | 1.24 | 11.98 |
| AAO3 | c33740.graph_c0 | 4559 | 22.27 | 18.46 | 7.99 | 2.5 | 19.43 | 6.51 | 1.22 | 22.02 | 5.97 |
| CYP707A1 | c20275.graph_c0 | 2041 | 2.11 | 2.97 | 7.5 | 25.59 | 4.14 | 27.64 | 85.48 | 5.58 | 13.47 |
| CYP707A3 | c27956.graph_c0 | 1811 | 3.92 | 5.03 | 2.61 | 1.36 | 4.26 | 2.13 | 1.11 | 3.72 | 0.46 |
| CYP707A4 | c35953.graph_c0 | 1734 | 17.32 | 8.77 | 1.16 | 1.22 | 9.11 | 0.89 | 0.81 | 11.71 | 0.59 |
| Gene ID | FPKM (max) | FPKM (min) | Description for the best hit in C. sinensis | Number of correlations |
|---|---|---|---|---|
| c11225.graph_c0 | 17.95 | 0.69 | bHLH107 | 4 |
| 12645.graph_c0 | 42.68 | 1.02 | BIM1 isoform X1 | 3 |
| c21405.graph_c0 | 3.45 | 0 | bHLH162 | 2 |
| c22086.graph_c0 | 9.37 | 0 | bHLH57 | 2 |
| c2272.graph_c0 | 2.60 | 0 | bHLH35 | 2 |
| c22910.graph_c0 | 14.15 | 0.46 | bHLH61 | 4 |
| c27501.graph_c0 | 21.44 | 5.78 | bHLH130 | 4 |
| c589.graph_c1 | 3.97 | 0 | HEC2 | 2 |
| c6149.graph_c0 | 2.74 | 0 | ILR3 | 4 |
| c34091.graph_c0 | 256.50 | 6.32 | Ethylene-responsive transcription factor ERF12 | 3 |
| c24274.graph_c0 | 12.57 | 1.31 | Ethylene-responsive transcription factor-like protein | 3 |
| c34297.graph_c0 | 138.23 | 3.30 | Ethylene-responsive transcription factor ERF107 | 3 |
| c407.graph_c1 | 3.13 | 0 | Ethylene-responsive transcription factor 13 | 2 |
| c8122.graph_c0 | 111.06 | 5.19 | Ethylene-responsive transcription factor RAP2-3 | 4 |
| c8622.graph_c0 | 57.79 | 1.26 | Ethylene-responsive transcription factor ERF23 | 2 |
| c30160.graph_c0 | 22.31 | 0.04 | AP2-like ethylene-responsive transcription factor ANT | 1 |
| c26695.graph_c0 | 2.83 | 0 | AP2-like ethylene-responsive transcription factor AIL1 | 4 |
| c30127.graph_c0 | 118.39 | 13.99 | Auxin response factor 4 | 3 |
| c27816.graph_c0 | 5.91 | 0 | Growth-regulating factor 1 | 3 |
| c9781.graph_c0 | 5.38 | 0 | Growth-regulating factor 4 | 2 |
| c22501.graph_c0 | 8.35 | 0.91 | Transcriptional adapter ADA2b isoform X1 | 6 |
| c23847.graph_c0 | 1296.81 | 106.33 | MYB1R1-like | 4 |
| c25031.graph_c0 | 2.34 | 0 | MYB11 | 4 |
| c9240.graph_c0 | 6.37 | 0.20 | MYB52 | 2 |
| c1320.graph_c0 | 4.49 | 0 | MYB13 | 2 |
| c13169.graph_c0 | 104.34 | 1.68 | Transcription factor AS1-like | 6 |
| c13009.graph_c0 | 157.18 | 2.18 | REVEILLE 1 | 2 |
| c21421.graph_c0 | 23.56 | 0.45 | Transcription factor AS1 | 6 |
| c13063.graph_c0 | 3047.44 | 242.76 | Mini zinc finger protein 2 | 4 |
| c21980.graph_c0 | 6.22 | 0.19 | WRKY22 | 4 |
| c34311.graph_c0 | 114.93 | 21.37 | WRKY17 | 5 |
| c13516.graph_c0 | 26.60 | 4.39 | NAC72 | 3 |
| c31393.graph_c0 | 20.63 | 3.05 | Transcription factor RF2a-like | 3 |
| c28763.graph_c0 | 23.77 | 0.83 | Transcription factor TGA7 | 3 |
| c34869.graph_c0 | 58.15 | 5.27 | BES1/BZR1 homolog protein 4 | 6 |
| c35277.graph_c0 | 22.80 | 7.03 | Transcription factor TCP20 | 3 |
| c35957.graph_c0 | 17.19 | 1.35 | Trihelix transcription factor GTL1 | 2 |
| c31746.graph_c0 | 330.33 | 15.64 | DELLA protein GAI | 5 |
| c31940.graph_c0 | 4.21 | 0 | FAR1-related sequence 5-like | 5 |
| c28566.graph_c0 | 13.07 | 1.05 | Transcription factor HHO2-like | 6 |
| c29740.graph_c0 | 26.66 | 1.14 | B3 domain-containing transcription factor VRN1-like | 4 |
| c27534.graph_c0 | 303.99 | 13.66 | Effector of transcription 2 | 4 |
| c27216.graph_c0 | 110.23 | 3.11 | Transcription factor TCP4 | 6 |
| c22856.graph_c0 | 7.47 | 0 | Trihelix transcription factor GT-2 | 4 |
| c20987.graph_c0 | 30.86 | 0.03 | GATA transcription factor 11 | 4 |
| c14734.graph_c0 | 63.08 | 6.70 | Transcription factor HHO3 | 5 |
| c12838.graph_c0 | 49.59 | 9.91 | YABBY 2 isoform X1 | 5 |
| c10930.graph_c0 | 22.79 | 1.61 | Trihelix transcription factor PTL | 6 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, C.-C.; Zhang, Y.-F.; Lin, Y.-J.; Chen, Y.; Lu, X.-K. Illumina® Sequencing Reveals Candidate Genes of Carotenoid Metabolism in Three Pummelo Cultivars (Citrus Maxima) with Different Pulp Color. Int. J. Mol. Sci. 2019, 20, 2246. https://doi.org/10.3390/ijms20092246
Jiang C-C, Zhang Y-F, Lin Y-J, Chen Y, Lu X-K. Illumina® Sequencing Reveals Candidate Genes of Carotenoid Metabolism in Three Pummelo Cultivars (Citrus Maxima) with Different Pulp Color. International Journal of Molecular Sciences. 2019; 20(9):2246. https://doi.org/10.3390/ijms20092246
Chicago/Turabian StyleJiang, Cui-Cui, Yan-Fang Zhang, Yan-Jin Lin, Yuan Chen, and Xin-Kun Lu. 2019. "Illumina® Sequencing Reveals Candidate Genes of Carotenoid Metabolism in Three Pummelo Cultivars (Citrus Maxima) with Different Pulp Color" International Journal of Molecular Sciences 20, no. 9: 2246. https://doi.org/10.3390/ijms20092246