Transcriptome Changes in the Mink Uterus during Blastocyst Dormancy and Reactivation
Abstract
:1. Introduction
2. Results
2.1. Identification of Transcriptomic Differences
2.2. Gene Ontology Analysis of the DEGs
2.3. KEGG Pathway Analysis of DEGs
2.4. PPI Network Analysis
2.5. Candidate Gene Selection and Validation of Expression Levels
2.6. Prolactin Activates Diapauses Embryo by PI3K Signaling Pathway
3. Discussion
4. Materials and Methods
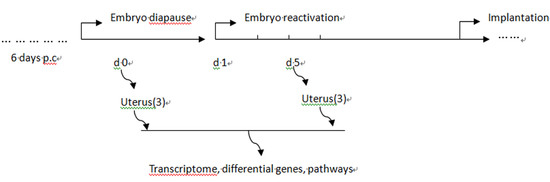
4.1. Animals and Sample Collection
4.2. RNA Sequencing
4.3. Enrichment Analysis of DEGs
4.4. Validation of DEGs
4.5. PPI (Protein Protein Interaction) Network Construction
4.6. Embryo Culture
4.7. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Enders, R.K. Reproduction in the mink (Mustela vison). Am. Philos. Soc. 1952, 96, 691–755. [Google Scholar]
- Fenelon, J.C.; Banerjee, A.; Lefevre, P.; Gratian, F.; Murphy, B.D. Polyamine-Mediated Effects of Prolactin Dictate Emergence from Mink Obligate Embryonic Diapause. Biol. Reprod. 2016, 95, 6. [Google Scholar] [CrossRef] [PubMed]
- Chang, M. Reciprocal insemination and egg transfer between ferrets and mink. Ecol. Genet. Physiol. 1968, 168, 49–59. [Google Scholar] [CrossRef]
- Moreau, G.M.; Arslan, A.; Douglas, D.A.; Song, J.; Smith, L.C.; Murphy, B.D. Development of immortalized endometrial epithelial and stromal cell lines from the mink (Mustela vison) uterus and their effects on the survival in vitro of mink blastocysts in obligate diapause. Biol. Reprod. 1995, 53, 511–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, J.H.; Houde, A.; Murphy, B.D. Cloning of leukemia inhibitory factor (LIF) and its expression in the uterus during embryonic diapause and implantation in the mink (Mustela vison). Mol. Reprod. Dev. 1998, 51, 13–21. [Google Scholar] [CrossRef]
- Desmarais, J.A.; Cao, M.; Bateman, A.; Murphy, B.D. Spatiotemporal expression pattern of progranulin in embryo implantation and placenta formation suggests a role in cell proliferation, remodeling, and angiogenesis. Reproduction 2008, 136, 247–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lefevre, P.L.; Palin, M.F.; Beaudry, D.; Dobias-Goff, M.; Desmarais, J.A.; Llerena, E.M.; Murphy, B.D. Uterine signaling at the emergence of the embryo from obligate diapause. Am. J. Physiol. Endocrinolog. Metab. 2011, 300, E800–E808. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; An, X.P.; Liu, X.R.; Fu, M.Z.; Han, P.; Peng, J.Y.; Hou, J.X.; Zhou, Z.Q.; Cao, B.Y.; Song, Y.X. Characterization of the Transcriptional Complexity of the Receptive and Pre-receptive Endometria of Dairy Goats. Sci. Rep. 2015, 5, 14244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samborski, A.; Graf, A.; Krebs, S.; Kessler, B.; Reichenbach, M.; Reichenbach, H.D.; Ulbrich, S.E.; Bauersachs, S. Transcriptome changes in the porcine endometrium during the preattachment phase. Biol. Reprod. 2013, 89, 134. [Google Scholar] [CrossRef]
- McRae, A.C. Observation on the Timing of Embryo Mortality in Ranch Mink (Mustela Vison); Canada Mink Breeders Association: Toronto, Canada, 1992; pp. 35–48. [Google Scholar]
- Fenelon, J.C.; Shaw, G.; Frankenberg, S.R.; Murphy, B.D.; Renfree, M.B. Embryo arrest and reactivation: potential candidates controlling embryonic diapause in the tammar wallaby and mink. Biol. Reprod. 2017, 96, 877–894. [Google Scholar] [CrossRef]
- Desmarais, J.A.; Bordignon, V.; Lopes, F.L.; Smith, L.C.; Murphy, B.D. The escape of the mink embryo from obligate diapause. Biol. Reprod. 2004, 70, 662–670. [Google Scholar] [CrossRef] [PubMed]
- Reese, J.; Binart, N.; Brown, N.; Ma, W.G.; Paria, B.C.; Das, S.K.; Kelly, P.A.; Dey, S.K. Implantation and decidualization defects in prolactin receptor (PRLR)-deficient mice are mediated by ovarian but not uterine PRLR. Endocrinology 2000, 141, 1872–1881. [Google Scholar] [CrossRef]
- Brockman, J.L.; Schroeder, M.D.; Schuler, L.A. PRL activates the cyclin D1 promoter via the Jak2/Stat pathway. Mol. Endocrinol. 2002, 16, 774–784. [Google Scholar] [CrossRef] [PubMed]
- Jabbour, H.N.; Critchley, H.O.; Boddy, S.C. Expression of functional prolactin receptors in nonpregnant human endometrium: Janus kinase-2, signal transducer and activator of transcription-1 (STAT1), and STAT5 proteins are phosphorylated after stimulation with prolactin. J. Clin. Endocrinol. Metab. 1998, 83, 2545–2553. [Google Scholar] [CrossRef]
- Berlanga, J.J.; Gualillo, O.; Buteau, H.; Applanat, M.; Kelly, P.A.; Edery, M. Prolactin activates tyrosyl phosphorylation of insulin receptor substrate 1 and phosphatidylinositol-3-OH kinase. J. Biol. Chem. 1997, 272, 2050–2052. [Google Scholar] [CrossRef] [PubMed]
- Bachelot, A.; Binart, N. Reproductive role of prolactin. Reproduction 2007, 133, 361–369. [Google Scholar] [CrossRef] [Green Version]
- Dai, J.; Peng, L.; Fan, K.; Wang, H.; Wei, R.; Ji, G.; Cai, J.; Lu, B.; Li, B.; Zhang, D.; et al. Osteopontin induces angiogenesis through activation of PI3K/AKT and ERK1/2 in endothelial cells. Oncogene 2009, 28, 3412–3422. [Google Scholar] [CrossRef]
- Karar, J.; Maity, A. PI3K/AKT/mTOR Pathway in Angiogenesis. Front. Mol. Neurosci. 2011, 4, 51. [Google Scholar] [CrossRef] [PubMed]
- Goncharova, E.A.; Ammit, A.J.; Irani, C.; Carroll, R.G.; Eszterhas, A.J.; Panettieri, R.A.; Krymskaya, V.P. PI3K is required for proliferation and migration of human pulmonary vascular smooth muscle cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 2002, 283, L354–L363. [Google Scholar] [CrossRef]
- Bakin, A.V.; Tomlinson, A.K.; Bhowmick, N.A.; Moses, H.L.; Arteaga, C.L. Phosphatidylinositol 3-kinase function is required for transforming growth factor beta-mediated epithelial to mesenchymal transition and cell migration. J. Biol. Chem. 2000, 275, 36803–36810. [Google Scholar] [CrossRef]
- Zeng, X.; Mao, X.; Huang, Z.; Wang, F.; Wu, G.; Qiao, S. Arginine enhances embryo implantation in rats through PI3K/PKB/mTOR/NO signaling pathway during early pregnancy. Reproduction 2013, 145, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, P.; Sapru, K.; Strakova, Z.; Fazleabas, A.T. Chorionic gonadotropin regulates prostaglandin E synthase via a phosphatidylinositol 3-kinase-extracellular regulatory kinase pathway in a human endometrial epithelial cell line: implications for endometrial responses for embryo implantation. Endocrinology 2009, 150, 4326–4337. [Google Scholar] [CrossRef]
- Gentilini, D.; Busacca, M.; Di Francesco, S.; Vignali, M.; Vigano, P.; Di Blasio, A.M. PI3K/Akt and ERK1/2 signalling pathways are involved in endometrial cell migration induced by 17beta-estradiol and growth factors. Mol. Hum. Reprod. 2007, 13, 317–322. [Google Scholar] [CrossRef] [PubMed]
- Mosavi, L.K.; Cammett, T.J.; Desrosiers, D.C.; Peng, Z.Y. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004, 13, 1435–1448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, H.; Wang, H.; Wang, Y.; Liu, C.; Wang, C.; Guo, J. Transcriptomic Analysis of the Porcine Endometrium during Embryo Implantation. Genes 2015, 6, 1330–1346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Garcia, J.; Kolp, L.; Cheadle, C.; Rodriguez, A.; Vlahos, N.F. The impact of luteal phase support on gene expression of extracellular matrix protein and adhesion molecules in the human endometrium during the window of implantation following controlled ovarian stimulation with a GnRH antagonist protocol. Fertil. Steril. 2010, 94, 2264–2271. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Yu, J.H.; Lee, M.Y.; Kim, A.L.; Jo, M.H.; Kim, M.; Cho, S.R.; Kim, Y.H. Differential Expression of Extracellular Matrix and Adhesion Molecules in Fetal-Origin Amniotic Epithelial Cells of Preeclamptic Pregnancy. PLoS ONE 2016, 11, e0156038. [Google Scholar] [CrossRef]
- Kong, S.; Han, X.; Cui, T.; Zhou, C.; Jiang, Y.; Zhang, H.; Wang, B.; Wang, H.; Zhang, S. MCM2 mediates progesterone-induced endometrial stromal cell proliferation and differentiation in mice. Endocrine 2016, 53, 595–606. [Google Scholar] [CrossRef]
- Ray, S.; Pollard, J.W. KLF15 negatively regulates estrogen-induced epithelial cell proliferation by inhibition of DNA replication licensing. Proc. Natl. Acad. Sci. USA 2012, 109, E1334–E1343. [Google Scholar] [CrossRef] [Green Version]
- Pilbeam, T.E.; Concannon, P.W.; Travis, H.F. The annual reproductive cycle of mink (Mustela vison). J. Anim. Sci. 1979, 48, 578–584. [Google Scholar] [CrossRef]
- Rozhnov, V.V.; Naidenko, S.V.; Naidenko, S.V. Variation of the level of steroid hormones in the blood plasma of three Mustelide Species (Mammalia, Carnivora, Mustelidae) during the annual cycle. Dokl. Biol. Sci. 2007, 413, 121–124. [Google Scholar] [CrossRef]
- Papke, R.L.; Concannon, P.W.; Travis, H.F.; Hansel, W. Control of luteal function and implantation in the mink by prolactin. J. Anim. Sci. 1980, 50, 1102–1107. [Google Scholar] [CrossRef] [PubMed]
- Murphy, B.D.; Concannon, P.W.; Travis, H.F.; Hansel, W. Prolactin: the hypophyseal factor that terminates embryonic diapause in mink. Biol. Reprod. 1981, 25, 487–491. [Google Scholar] [CrossRef] [PubMed]
- Murphy, B.D.; Mead, R.A.; McKibbin, P.E. Luteal contribution to the termination of preimplantation delay in mink. Biol. Reprod. 1983, 28, 497–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spiess, K.; Teodoro, W.R.; Zorn, T.M. Distribution of collagen types I, III, and V in pregnant mouse endometrium. Connect. Tissue Res. 2007, 48, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Vidal-Vanaclocha, F.; Alvarez, A.; Asumendi, A.; Urcelay, B.; Tonino, P.; Dinarello, C.A. Interleukin 1 (IL-1)-dependent melanoma hepatic metastasis in vivo; increased endothelial adherence by IL-1-induced mannose receptors and growth factor production in vitro. J. Natl. Cancer Inst. 1996, 88, 198–205. [Google Scholar] [CrossRef]
- Apte, R.N.; Voronov, E. Is interleukin-1 a good or bad ‘guy’ in tumor immunobiology and immunotherapy? Immunological 2008, 222, 222–241. [Google Scholar] [CrossRef]
- Meager, A. Cytokine regulation of cellular adhesion molecule expression in inflammation. Cytokine Growth Factor Rev. 1999, 10, 27–39. [Google Scholar] [CrossRef]
- Sheth, K.V.; Roca, G.L.; al-Sedairy, S.T.; Parhar, R.S.; Hamilton, C.J.; al-Abdul Jabbar, F. Prediction of successful embryo implantation by measuring interleukin-1-alpha and immunosuppressive factor(s) in preimplantation embryo culture fluid. Fertil. Steril. 1991, 55, 952–957. [Google Scholar] [CrossRef]
- Simon, C.; Valbuena, D.; Krussel, J.; Bernal, A.; Murphy, C.R.; Shaw, T.; Pellicer, A.; Polan, M.L. Interleukin-1 receptor antagonist prevents embryonic implantation by a direct effect on the endometrial epithelium. Fertil. Steril. 1998, 70, 896–906. [Google Scholar] [CrossRef]
- Tanikawa, M.; Lee, H.Y.; Watanabe, K.; Majewska, M.; Skarzynski, D.J.; Park, S.B.; Lee, D.S.; Park, C.K.; Acosta, T.J.; Okuda, K. Regulation of prostaglandin biosynthesis by interleukin-1 in cultured bovine endometrial cells. J. Endocrinol. 2008, 199, 425–434. [Google Scholar] [CrossRef] [Green Version]
- Majewska, M.; Woclawek-Potocka, I.; Bah, M.M.; Hapunik, J.; Piotrowska, K.K.; Tasaki, Y.; Acosta, T.J.; Okuda, K.; Skarzynski, D.J. Is interleukin-1α a luteotrophic or luteolytic agent in cattle? Reproduction 2010, 139, 665–672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kennedy, T.G. Timing of uterine sensitivity for the decidual cell reaction: role of prostaglandins. Biol. Reprod. 1980, 22, 519–525. [Google Scholar] [CrossRef]
- Yee, G.M.; Kennedy, T.G. Prostaglandin E2, cAMP and cAMP-dependent protein kinase isozymes during decidualization of rat endometrial stromal cells in vitro. Prostaglandins 1993, 46, 117–138. [Google Scholar] [CrossRef]
- Han, S.W.; Lei, Z.M.; Rao, C.V. Up-regulation of cyclooxygenase-2 gene expression by chorionic gonadotropin during the differentiation of human endometrial stromal cells into decidua. Endocrinology 1996, 137, 1791–1797. [Google Scholar] [CrossRef] [PubMed]
- Simon, C.; Martin, J.C.; Pellicer, A. Paracrine regulators of implantation. Best Pract. Res. Clin. Obstet. Gynaecol. 2000, 14, 815–826. [Google Scholar] [CrossRef] [PubMed]
- Guzeloglu-Kayisli, O.; Kayisli, U.A.; Taylor, H.S. The role of growth factors and cytokines during implantation: endocrine and paracrine interactions. Semin. Reprod. Med. 2009, 27, 62–79. [Google Scholar] [CrossRef] [PubMed]
- Clementi, C.; Tripurani, S.K.; Large, M.J.; Edson, M.A.; Creighton, C.J.; Hawkins, S.M.; Kovanci, E.; Kaartinen, V.; Lydon, J.P.; Pangas, S.A.; et al. Activin-like kinase 2 functions in peri-implantation uterine signaling in mice and humans. PLoS Genet. 2013, 9, e1003863. [Google Scholar] [CrossRef] [PubMed]
- Todt, J.C.; Yang, Y.; Lei, J.; Lauria, M.R.; Sorokin, Y.; Cotton, D.B.; Yelian, F.D. Effects of tumor necrosis factor-alpha on human trophoblast cell adhesion and motility. Am. J. Reprod. Immunol. 1996, 36, 65–71. [Google Scholar] [CrossRef]
- Keck, P.J.; Hauser, S.D.; Krivi, G.; Sanzo, K.; Warren, T.; Feder, J.; Connolly, D.T. Vascular permeability factor, an endothelial cell mitogen related to PDGF. Science 1989, 246, 1309–1312. [Google Scholar] [CrossRef] [PubMed]
- Lopes, F.L.; Desmarais, J.; Gevry, N.Y.; Ledoux, S.; Murphy, B.D. Expression of vascular endothelial growth factor isoforms and receptors Flt-1 and KDR during the peri-implantation period in the mink, Mustela vison. Biol. Reprod. 2003, 68, 1926–1933. [Google Scholar] [CrossRef] [PubMed]
- Aghajanova, L.; Bjuresten, K.; Altmae, S.; Landgren, B.M.; Stavreus-Evers, A. HB-EGF but not amphiregulin or their receptors HER1 and HER4 is altered in endometrium of women with unexplained infertility. Reprod. Sci. 2008, 15, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Francis, J.; Rai, R.; Sebire, N.J.; El-Gaddal, S.; Fernandes, M.S.; Jindal, P.; Lokugamage, A.; Regan, L.; Brosens, J.J. Impaired expression of endometrial differentiation markers and complement regulatory proteins in patients with recurrent pregnancy loss associated with antiphospholipid syndrome. Mol. Hum. Reprod. 2006, 12, 435–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, S.L.; Lessey, B.A.; Fritz, M.A.; Meyer, W.R.; Murray, M.J.; Speckman, P.L.; Nowicki, B.J. In vivo and in vitro evidence suggest that HB-EGF regulates endometrial expression of human decay-accelerating factor. J. Clin. Endocrinol. Metab. 2002, 87, 1368–1375. [Google Scholar] [CrossRef] [PubMed]
- Ohtsuki, T.; Otsuki, M.; Murakami, Y.; Hirata, K.; Takeuchi, S.; Takahashi, S. Alternative leader-exon usage in mouse IGF-I mRNA variants: class 1 and class 2 IGF-I mRNAs. Zool. Sci. 2007, 24, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Diane, M.K.; Hewitt, S.C.; Ciana, P.; Raviscioni, M.; Lindzey, J.K.; Foley, J.; Maggi, A.; DiAugustine, R.P.; Korach, K.S. Requirement of estrogen receptor-α in insulin-like growth factor-1 (IGF-1)-induced uterine responses and in vivo evidence for IGF-1/estrogen receptor cross-talk. J. Biol. Chem. 2002, 277, 8531–8537. [Google Scholar]
- Gerber, R.T.; Anwar, M.A.; Poston, L. Enhanced acetylcholine induced relaxation in small mesenteric arteries from pregnant rats: an important role for endothelium-derived hyperpolarizing factor (EDHF). Br. J. Pharmacol. 1998, 125, 455–460. [Google Scholar] [CrossRef] [Green Version]
- Jiang, B.H.; Zheng, J.Z.; Aoki, M.; Vogt, P.K. Phosphatidylinositol 3-kinase signaling mediates angiogenesis and expression of vascular endothelial growth factor in endothelial cells. Proc. Natl. Acad. Sci. USA 2000, 97, 1749–1753. [Google Scholar] [CrossRef] [Green Version]
- Salker, M.S.; Steel, J.H.; Hosseinzadeh, Z.; Nautiyal, J.; Webster, Z.; Singh, Y.; Brucker, S.; Lang, F.; Brosens, J.J. Activation of SGK1 in Endometrial Epithelial Cells in Response to PI3K/AKT Inhibition Impairs Embryo Implantation. Cell. Physiol. Biochem. 2016, 39, 2077–2087. [Google Scholar] [CrossRef] [Green Version]
- Graupera, M.; Guillermet-Guibert, J.; Foukas, L.C.; Phng, L.K.; Cain, R.J.; Salpekar, A.; Pearce, W.; Meek, S.; Millan, J.; Cutillas, P.R.; et al. Angiogenesis selectively requires the p110alpha isoform of PI3K to control endothelial cell migration. Nature 2008, 453, 662–666. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.D.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Cao, X.; Xu, C.; Zhang, Y.; Wei, H.; Liu, Y.; Cao, J.; Zhao, W.; Bao, K.; Wu, Q. Comparative transcriptome analysis of embryo invasion in the mink uterus. Placenta 2019, 75, 16–22. [Google Scholar] [CrossRef]






| Comparison a | Fold > 2 b | Fold > 1.5 c | p < 0.05 d |
|---|---|---|---|
| Activated > Diapause | 622 (738) | 167 (178) | 963 (1063) |
| Diapause > Activated | 445 (502) | 58 (67) | 564 (621) |
| All | 1067 (1240) | 225 (245) | 1527 (1684) |
| Gene Symbol | Log2 Fold Change | p Value | Gene Description |
|---|---|---|---|
| ANKS1A | 7.9822 | 6.52 × 10−42 | ankyrin repeat and SAM domain-containing protein 1A isoform X2 |
| TEAD | −7.7671 | 3.41 × 10−37 | Tead3 protein |
| LAMA3 | 7.4272 | 2.01 × 10−33 | laminin, alpha 3 |
| XPO5 | −7.0783 | 8.76 × 10−31 | XPO5 protein |
| PNKD | 6.9436 | 1.04 × 10−27 | probable hydrolase PNKD isoform X2 |
| N4BP1 | −6.5331 | 3.17 × 10−34 | NEDD4-binding protein 1 |
| PDPR | 6.4222 | 6.67 × 10−29 | pyruvate dehydrogenase phosphatase regulatory subunit |
| CPSF3 | 6.3617 | 4.02 × 10−21 | cleavage and polyadenylation specific factor 3 |
| RIOK3 | 6.3151 | 6.04 × 10−23 | RIO kinase 3 |
| THOC1 | 6.2389 | 3.67 × 10−22 | THO complex 1 |
| RP11-489N6 | 6.1613 | 4.70 × 10−21 | BAC RP11-489N6 |
| LOC101694508 | −6.037 | 1.99 × 10−19 | uncharacterized LOC101694508 |
| DSG2 | 6.033 | 3.31 × 10−28 | desmoglein 2 |
| SLC7A2 | 6.0189 | 2.64 × 10−17 | solute carrier family 7 (cationic amino acid transporter, y) |
| ASPM, ASP | 5.9385 | 6.62 × 10−18 | asp (abnormal spindle) homolog, microcephaly associated |
| ATP6N | 5.9165 | 1.29 × 10−18 | V-type proton ATPase |
| ZXDC | 5.9001 | 3.30 × 10−19 | ZXD family zinc finger C |
| PDIA6 | 5.6566 | 6.67 × 10−17 | disulfide-isomerase A6 |
| PINK1 | −5.6508 | 2.77 × 10−17 | serine/threonine-protein kinase PINK1, mitochondrial |
| ERCC5 | −6.0343 | 6.47 × 10−20 | immunoglobulin-like variable motif containing (BIVM) |
| Category | Terms | p Value | Genes |
|---|---|---|---|
| BP | Xenobiotic metabolic process | 2.98×10−5 | ITGB4↓, ACP2↓, LAMA3_5↑, ADAMTS9↑, PTPRF↑, COL12A1↑, TN↓, CHL1↑, NCAM↑, EPHB6↑, serB↑, LIFR↓, PRLR↑, PSPH↑, IGDCC4↓, CSF2RB↑ |
| Cell communication | 6.38×10−5 | RASL11B↑, SLC18A2↑, HDHD2↑, ANK↓, IKBIP↑, NEMF↓, DNMBP↓, ARHGAP12↑, LOXL2_3_4↓, ATP13A1↑, TN↓, IGF1↑, BBC3↓, STAC↑, PKN↑, ANPRA↓, SYTL↑, IL1A↑, IFT27↓, CCRL1↑, PSD4↑, RASA2, CD44↓, NOTCH4↑, ARL4A↓, RCAN1↑, ELAC2↓, DDIT4L↓, BIVM↓, PLCD1↑, ZDHHC21↑, CXCL14↓, KIAA1324↑, LAMA1↑………. | |
| Signal transduction | 7.08×10−5 | SPAG5↑, TNFSF18↑, ARHGAP6↓, LCP1↑, SIPA1L2↑, ITGB4↓, VIPR2↑, LGR5↓, NAV1↓, SYT4↑, THOC1↑, ZYX↑, DMBT↓, PIKFYVE↓, LRP2↑, RAB36↑, RASAL1↓, COL12A1↑, EPHB6↑, SPOCK↓, PPP2R3↑, STAT5A↓, NR4A1↑, CSF2RB↑, SIRPA↓……. | |
| Cellular response to stimulus | 9.02×10−5 | PDE7↑, NFKBIZ↓, PRSS15↓, KIAA1324↑, IGDCC4↓, PDZK1↑, TTC39A↑, RFX1_2_3↓, SEPT3_9_12↑, SLC41A↑, KIAA1324↑, CASR↓, GDF3↑, PLCB↑, NOD2↓, VEGFA↑, STAT2↑, PDE1↑, GPR143↑, ADCYAP1R1↓, THOC1↑, IGSF10↓, EGF↑, PDZD8↑……. | |
| CC | Extracellular matrix | 7.05×10−9 | COL1AS↑, ATRNL1↑, COL4A↑, ADAMTS9↑, GPC6↑, LCP1↑, SPARC↑, PLS1↑, PPP2R3↑, SPOCK↓, SPTAN1↑, COL12A1↑, COL6A↓, ADAM33↓, PLS1↑, PLCD↑, SPARC↑, ADAMTS12↑, COL1AS↑, |
| Collagen trimer | 7.79×10−5 | COL1AS↑, COL4A↑ | |
| MF | Phosphatase activity | 8.98×10−6 | PTPRF↑, ARL4↓, ALCAM↑, ACP2↓, EPHB6↑, CPZ↑, NCAM↑, COL12A1↑, PTP4A↑, SERB↑, CHL1↑, PHOSPHO2↑, EPHB6↑, DUSP↑, PTPRR↑, PRLR↑, CSF2RB↑, ADAMTS9↑, CCBL↑, ITGB4↓, DCHS1_2↑ |
| N,N-dimethylaniline monooxygenase activity | 4.28×10−4 | FMO↑, IL4I1↑ | |
| G-protein coupled receptor activity | 4.42×10−4 | GPR143↑, GPR161↓, PACAPRI↓, TES↑, CNR1↑, VIPR2↑, MRGPRF↓, TMEM161B↑, NSUN2↑, CCRL1↑, PDZK1↑, FZD5_8↑, CASR↓, LGR5↓, KIAA1324↑, ADRA2C↓, RGS3↑, PAMR1↓, PDZD8↑, LGR5↓, HRH1↓ | |
| Metallocarboxypeptidase activity | 7.08×10−4 | CPXM1↑, CPZ↑, CPN1↑, CPXM2↑ |
| KEGG Pathway | p Value | Genes |
|---|---|---|
| ECM-receptor interaction | 1.61×10−11 | COL1AS↑, CD44↓, LAMA1_2↑, TN↓, LAMA3_5↑, ITGB4↓, LAMC3↑, THBS1↓, COL6A↓, ITGA5↓, ITGB8↓, ITGA9↓, COL4A↑, CHAD↑, THBS2S↓ |
| Focal adhesion | 7.53×10−08 | PIK3C↑, COL1AS↑, LAMA1_2↑, TN↓, LAMA3_5↑, FLNA↓, EGF↑, ITGB4↓, ROCK2↑, VEGFA↑, THBS1↓, PDPK1↑, ACTB_G1↑, COL6A↓, IGF1↑, ITGA5↓, AKT↑, ITGB8↓, LAMC3↑, PPP1C↑, ITGA9↓, COL4A↑, CHAD↑, THBS2S↓, CCND1↓ |
| PI3K-Akt signaling pathway | 4.94×10−7 | PIK3C↑, COL1AS↑, LAMA1_2↑, COL3A1↑, TN↓, LAMA3_5↑, VEGFA↑, PPP2R3↑, MDM2↓, EGF↑, ITGB4↓, PRKAA↓, THBS1↓, PRLR↑, PKN↑, COL6A↓, IGF1↑, ITGA5↓, AKT↑, ITGB8↓, PDPK1↑, YWHAB_Q_Z↑, NR4A1↑, LAMC3↑, PHLPP↑, ITGA9↓, COL4A↑, CHAD↑, SGK1↑, THBS2S↓, FGFR4↑, CCND1↓ |
| Cytokine-cytokine receptor interaction | 2.37×10−2 | VEGFA↑, TNFSF12↓, IL13RA1↓, CXCL14↓, CSF2RB↑, PRLR↑, IL1A↑, EGF↑, TNFSF18↑, LIFR↓, LTBR↓, BMPR1B↑ |
| Protein digestion and absorption | 7.65×10−6 | COL1AS↑, ACE2↑, XPNPEP2↑, COL6A↓, SLC7A9↓, COL4A↑, SLC15A1↑, ACEH↑ |
| AGE-RAGE signaling pathway in diabetic complications | 6.35×10−5 | PIK3C↑, COL1AS↑, PLCB↑, VEGFA↑, PLCD↑, IL1A↑, STAT5A↓, AKT↑, COL4A↑, CCND1↓ |
| Cell adhesion molecules (CAMs) | 1.69×10−3 | MHC2↓, CLDN↑, ITGB8↓, ALCAM↑, CDH4↑, NCAM↑, PTPRF↑, SIGLEC1↓, ITGAM↓, ITGA9↓, PTPRC↑, NRCAM↑ |
| Glycine, serine and threonine metabolism | 3.54×10−3 | GATM↑, serB↑, SARDH↑, CTH↑, gcvT↑, DAO↑, DLD↑ |
| Thyroid hormone synthesis | 6.34×10−3 | PLCB↑, GPX8↑, TTF2↑, SLC26A4↑, LRP2↑, ADCY7↓, PAX8↑ |
| Platelet activation | 0.92×10−2 | ROCK2↑, COL1AS↑, AKT↑, PIK3C↑, PLCB↑, PPP1C↑, ACTB_G1↑, ADCY7↓ |
| Prolactin signaling pathway | 0.50×10−2 | ELF5↑, PIK3C↑, AKT↑, CYP17A↑, PRLR↑, STAT5A↓, CCND1↓ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, X.; Zhao, J.; Liu, Y.; Ba, H.; Wei, H.; Zhang, Y.; Wang, G.; Murphy, B.D.; Xing, X. Transcriptome Changes in the Mink Uterus during Blastocyst Dormancy and Reactivation. Int. J. Mol. Sci. 2019, 20, 2099. https://doi.org/10.3390/ijms20092099
Cao X, Zhao J, Liu Y, Ba H, Wei H, Zhang Y, Wang G, Murphy BD, Xing X. Transcriptome Changes in the Mink Uterus during Blastocyst Dormancy and Reactivation. International Journal of Molecular Sciences. 2019; 20(9):2099. https://doi.org/10.3390/ijms20092099
Chicago/Turabian StyleCao, Xinyan, Jiaping Zhao, Yong Liu, Hengxing Ba, Haijun Wei, Yufei Zhang, Guiwu Wang, Bruce D. Murphy, and Xiumei Xing. 2019. "Transcriptome Changes in the Mink Uterus during Blastocyst Dormancy and Reactivation" International Journal of Molecular Sciences 20, no. 9: 2099. https://doi.org/10.3390/ijms20092099