TET1 Knockdown Inhibits Porphyromonas gingivalis LPS/IFN-γ-Induced M1 Macrophage Polarization through the NF-κB Pathway in THP-1 Cells
Abstract
:1. Introduction
2. Results
2.1. Pg. LPS/IFN-γ Polarized M0 Macrophages into M1 Proinflammatory Macrophages
2.2. TET1 Expression Decreased in LPS/IFN-γ-Induced M1 Macrophages
2.3. TET1 Knockdown Inhibited the Expression of Proinflammatory Cytokines and Chemokines during LPS/IFN-γ-Induced M1 Macrophage Activation
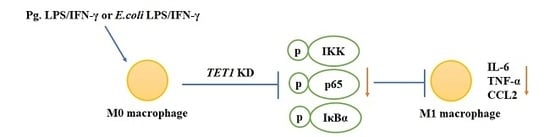
2.4. TET1 Knockdown Attenuated M1 Macrophage Polarization through the NF-κB Signaling Pathway
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. M1 Macrophage Polarization
4.3. Cell Counting Kit-8 (CCK8) Assay
4.4. TET1 Small Interfering RNA (siRNA) Transfection
4.5. Real-Time Quantitative Polymerase Chain Reaction (qRT-PCR)
4.6. Western Blot Analysis
4.7. Enzyme-Linked Immunosorbent Assay (ELISA)
4.8. Fluorescence-Activated Cell Sorting Analysis (FACS)
4.9. Statistical Analysis
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| TET1 | ten-eleven translocation methyl cytosine dioxygenase 1 |
| IL-6 | Interleukin-6 |
| TNF-α | Tumor necrosis factor α |
| CCL2 | Chemokine ligand 2 |
| CCR7 | Chemokine receptor 7 |
| GAPDH | D-glyceraldehyde-3-phosphate dehydrogenase |
| Pg | Porphyromonas gingivalis |
| E. coli | Escherichia coli |
| LPS | Lipopolysaccharide |
| IFN-γ | Interferon-γ |
| ATCC | American type culture collection |
References
- An, T.; He, Q.; Xia, Y.; Chen, S.; Baral, S.; Mao, L.; Jin, H.; Li, Y.; Wang, M.; Chen, J.; et al. MiR-181b Antagonizes Atherosclerotic Plaque Vulnerability Through Modulating Macrophage Polarization by Directly Targeting Notch1. Mol. Neurobiol. 2017, 54, 6329–6341. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Bi, C.; Gao, L.; An, Y.; Chen, F.; Chen, F. Macrophage polarization in human gingival tissue in response to periodontal disease. Oral Dis. 2018, 25, 265–273. [Google Scholar] [CrossRef]
- Xu, R.; Zeng, G.; Wang, S.; Tao, H.; Ren, L.; Zhang, Z.; Zhang, Q.; Zhao, J.; Gao, J.; Li, D. Periodontitis promotes the diabetic development of obese rat via miR-147 induced classical macrophage activation. Biomed. Pharmacother. 2016, 83, 892–897. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Lin, C.; Xu, P.; Wu, S.; Fu, X.; Xia, W.; Yao, M. AGEs Induced Autophagy Impairs Cutaneous Wound Healing via Stimulating Macrophage Polarization to M1 in Diabetes. Sci. Rep. 2016, 6, 36416. [Google Scholar] [CrossRef]
- Lee, M.K.S.; Moore, X.; Fu, Y.; Al-Sharea, A.; Dragoljevic, D.; Fernandez-Rojo, M.A.; Parton, R.; Sviridov, D.; Murphy, A.J.; Chin-Dusting, J.P.F. High-density lipoprotein inhibits human M1 macrophage polarization through redistribution of caveolin-1. Br. J. Pharmacol. 2016, 173, 741–751. [Google Scholar] [CrossRef]
- Piccolo, V.; Curina, A.; Genua, M.; Ghisletti, S.; Simonatto, M.; Sabò, A.; Amati, B.; Ostuni, R.; Natoli, G. Opposing macrophage polarization programs show extensive epigenomic and transcriptional cross-talk. Nat. Immunol. 2017, 18, 530–540. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Holdbrooks, A.T.; Liu, Y.; Reynolds, S.L.; Yanagisawa, L.L.; Benveniste, E.N. SOCS3 deficiency promotes M1 macrophage polarization and inflammation. J. Immunol. 2012, 189, 3439–3448. [Google Scholar] [CrossRef]
- Wang, W.; Yuan, H.; Liu, G.; Ni, W.; Wang, F.; Tai, G. Escherichia coli Maltose-Binding Protein Induces M1 Polarity of RAW264.7 Macrophage Cells via a TLR2- and TLR4-Dependent Manner. Int. J. Mol. Sci. 2015, 16, 9896–9909. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, M.; Saqib, U.; Naim, A.; Roy, A.; Liu, D.; Bhatnagar, D.; Ravinder, R.; Baig, M.S. The TLR4-NOS1-AP1 signaling axis regulates macrophage polarization. Inflamm. Res. 2017, 66, 323–334. [Google Scholar] [CrossRef]
- Lin, S.; Chuang, H.; Ho, J.; Lee, C.; Hsiao, C. Treatment with TNF-alpha inhibitor rectifies M1 macrophage polarization from blood CD14+ monocytes in patients with psoriasis independent of STAT1 and IRF-1 activation. J. Dermatol. Sci. 2018, 91, 276–284. [Google Scholar] [CrossRef]
- Jiao, Y.; Hasegawa, M.; Inohara, N. Emerging roles of immunostimulatory oral bacteria in periodontitis development. Trends Microbiol. 2014, 22, 157–163. [Google Scholar] [CrossRef]
- Baek, K.J.; Ji, S.; Kim, Y.C.; Choi, Y. Association of the invasion ability of Porphyromonas gingivalis with the severity of periodontitis. Virulence 2015, 6, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Palm, E.; Demirel, I.; Bengtsson, T.; Khalaf, H. The role of toll-like and protease-activated receptors in the expression of cytokines by gingival fibroblasts stimulated with the periodontal pathogen Porphyromonas gingivalis. Cytokine 2015, 76, 424–432. [Google Scholar] [CrossRef] [PubMed]
- Ke, X.; Lei, L.; Li, H.; Li, H.; Yan, F. Manipulation of necroptosis by Porphyromonas gingivalis in periodontitis development. Mol. Immunol. 2016, 77, 8–13. [Google Scholar] [CrossRef]
- Ma, Z.; Dagnaes-Hansen, F.; Lovschall, H.; Song, W.; Nielsen, G.K.; Yang, C.; Wang, Q.; Kjems, J.; Gao, S. Macrophage-mediated nanoparticle delivery to the periodontal lesions in established murine model via Pg-LPS induction. J. Oral Pathol. Med. 2015, 44, 538–542. [Google Scholar] [CrossRef]
- Yang, J.; Zhu, Y.; Duan, D.; Wang, P.; Xin, Y.; Bai, L.; Liu, Y.; Xu, Y. Enhanced activity of macrophage M1/M2 phenotypes in periodontitis. Arch. Oral Biol. 2018, 96, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Lew, J.; Naruishi, K.; Kajiura, Y.; Nishikawa, Y.; Ikuta, T.; Kido, J.; Nagata, T. High Glucose-Mediated Cytokine Regulation in Gingival Fibroblasts and THP-1 Macrophage: A Possible Mechanism of Severe Periodontitis with Diabetes. Cell. Physiol. Biochem. 2018, 50, 973–986. [Google Scholar] [CrossRef]
- Lyko, F. The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 2018, 19, 81–92. [Google Scholar] [CrossRef]
- Ko, M.; An, J.; Pastor, W.A.; Koralov, S.B.; Rajewsky, K.; Rao, A. TET proteins and 5-methylcytosine oxidation in hematological cancers. Immunol. Rev. 2015, 263, 6–21. [Google Scholar] [CrossRef]
- Neves-Costa, A.; Moita, L.F. TET1 is a negative transcriptional regulator of IL-1beta in the THP-1 cell line. Mol. Immunol. 2013, 54, 264–270. [Google Scholar] [CrossRef]
- Yang, R.; Qu, C.; Zhou, Y.; Konkel, J.E.; Shi, S.; Liu, Y.; Chen, C.; Liu, S.; Liu, D.; Chen, Y.; et al. Hydrogen Sulfide Promotes Tet1- and Tet2-Mediated Foxp3 Demethylation to Drive Regulatory T Cell Differentiation and Maintain Immune Homeostasis. Immunity 2015, 43, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Feng, Z.; Li, Q.; Yi, B.; Xu, Q. DNA methylcytosine dioxygenase ten-eleven translocation 2 enhances lipopolysaccharide-induced cytokine expression in human dental pulp cells by regulating MyD88 hydroxymethylation. Cell Tissue Res. 2018, 373, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Genin, M.; Clement, F.; Fattaccioli, A.; Raes, M.; Michiels, C. M1 and M2 macrophages derived from THP-1 cells differentially modulate the response of cancer cells to etoposide. BMC Cancer 2015, 15, 577. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, J.; Peng, X.; Lv, B.; Wang, P.; Zhao, X.; Yu, B. Geraniin Inhibits LPS-Induced THP-1 Macrophages Switching to M1 Phenotype via SOCS1/NF-kappaB Pathway. Inflammation 2016, 39, 1421–1433. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Zhao, L.; Huang, X.; Ma, C.; Wang, Y.; Zhang, J.; Xuan, D. Enhanced Activity of the Macrophage M1/M2 Phenotypes and Phenotypic Switch to M1 in Periodontal Infection. J. Periodontol. 2016, 87, 1092–1102. [Google Scholar] [CrossRef]
- Van Delft, M.A.; Huitema, L.F.; Tas, S.W. The contribution of NF-kappaB signalling to immune regulation and tolerance. Eur. J. Clin. Investig. 2015, 45, 529–539. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Gaynor, R.B. IkappaB kinases: Key regulators of the NF-kappaB pathway. Trends Biochem. Sci. 2004, 29, 72–79. [Google Scholar] [CrossRef]
- Pires, B.; Silva, R.; Ferreira, G.M.; Abdelhay, E. NF-kappaB: Two Sides of the Same Coin. Genes (Basel) 2018, 9, 24. [Google Scholar] [CrossRef]
- Zuza, E.P.; Garcia, V.G.; Theodoro, L.H.; Ervolino, E.; Favero, L.F.V.; Longo, M.; Ribeiro, F.S.; Martins, A.T.; Spolidorio, L.C.; Zuanon, J.A.S.; et al. Influence of obesity on experimental periodontitis in rats: Histopathological, histometric and immunohistochemical study. Clin. Oral Invest. 2018, 22, 1197–1208. [Google Scholar] [CrossRef]
- Nagashima, H.; Shinoda, M.; Honda, K.; Kamio, N.; Watanabe, M.; Suzuki, T.; Sugano, N.; Sato, S.; Iwata, K. CXCR4 signaling in macrophages contributes to periodontal mechanical hypersensitivity in Porphyromonas gingivalis-induced periodontitis in mice. Mol. Pain. 2017, 13, 1–8. [Google Scholar] [CrossRef]
- Huang, X.; Yu, T.; Ma, C.; Wang, Y.; Xie, B.; Xuan, D.; Zhang, J. Macrophages Play a Key Role in the Obesity-Induced Periodontal Innate Immune Dysfunction via Nucleotide-Binding Oligomerization Domain-Like Receptor Protein 3 Pathway. J. Periodontol. 2016, 87, 1195–1205. [Google Scholar] [CrossRef]
- Su, L.; Zhang, W.; Wu, X.; Zhang, Y.; Chen, X.; Liu, G.; Chen, G.; Jiang, M. Glycocalyx-Mimicking Nanoparticles for Stimulation and Polarization of Macrophages via Specific Interactions. Small 2015, 11, 4191–4200. [Google Scholar] [CrossRef]
- Hou, Y.; Yu, H.; Liu, X.; Li, G.; Pan, J.; Zheng, C.; Yu, W. Gingipain of Porphyromonas gingivalis manipulates M1 macrophage polarization through C5a pathway. In Vitro Cell. Dev. Biol. Anim. 2017, 53, 593–603. [Google Scholar] [CrossRef]
- Talari, M.; Kapadia, B.; Kain, V.; Seshadri, S.; Prajapati, B.; Rajput, P.; Misra, P.; Parsa, K.V.L. MicroRNA-16 modulates macrophage polarization leading to improved insulin sensitivity in myoblasts. Biochimie 2015, 119, 16–26. [Google Scholar] [CrossRef]
- Lam, R.S.; O’Brien-Simpson, N.M.; Lenzo, J.C.; Holden, J.A.; Brammar, G.C.; Walsh, K.A.; McNaughtan, J.E.; Rowler, D.K.; Van Rooijen, N.; Reynolds, E.C. Macrophage depletion abates Porphyromonas gingivalis-induced alveolar bone resorption in mice. J. Immunol. 2014, 193, 2349–2362. [Google Scholar] [CrossRef]
- Gonzalez, O.A.; Novak, M.J.; Kirakodu, S.; Stromberg, A.; Nagarajan, R.; Huang, C.B.; Chen, K.C.; Orraca, L.; Martinez-Gonzalez, J.; Ebersole, J.L. Differential Gene Expression Profiles Reflecting Macrophage Polarization in Aging and Periodontitis Gingival Tissues. Immunol. Investig. 2015, 44, 643–664. [Google Scholar] [CrossRef]
- Lyu, J.; Bian, T.; Chen, B.; Cui, D.; Li, L.; Gong, L.; Yan, F. beta-defensin 3 modulates macrophage activation and orientation during acute inflammatory response to Porphyromonas gingivalis lipopolysaccharide. Cytokine 2017, 92, 48–54. [Google Scholar] [CrossRef]
- Cui, D.; Lyu, J.; Li, H.; Lei, L.; Bian, T.; Li, L.; Yan, F. Human beta-defensin 3 inhibits periodontitis development by suppressing inflammatory responses in macrophages. Mol. Immunol. 2017, 91, 65–74. [Google Scholar] [CrossRef]
- Neele, A.E.; Van den Bossche, J.; Hoeksema, M.A.; de Winther, M.P.J. Epigenetic pathways in macrophages emerge as novel targets in atherosclerosis. Eur. J. Pharmacol. 2015, 763, 79–89. [Google Scholar] [CrossRef]
- Cao, Q.; Wang, X.; Jia, L.; Mondal, A.K.; Diallo, A.; Hawkins, G.A.; Das, S.K.; Parks, J.S.; Yu, L.; Shi, H.; et al. Inhibiting DNA Methylation by 5-Aza-2’-deoxycytidine ameliorates atherosclerosis through suppressing macrophage inflammation. Endocrinology 2014, 155, 4925–4938. [Google Scholar] [CrossRef]
- Thangavel, J.; Samanta, S.; Rajasingh, S.; Barani, B.; Xuan, Y.; Dawn, B.; Rajasingh, J. Epigenetic modifiers reduce inflammation and modulate macrophage phenotype during endotoxemia-induced acute lung injury. J. Cell Sci. 2015, 128, 3094–3105. [Google Scholar] [CrossRef] [PubMed]
- Orlanski, S.; Labi, V.; Reizel, Y.; Spiro, A.; Lichtenstein, M.; Levin-Klein, R.; Koralov, S.B.; Skversky, Y.; Rajewsky, K.; Cedar, H.; et al. Tissue-specific DNA demethylation is required for proper B-cell differentiation and function. Proc. Natl. Acad. Sci. USA 2016, 113, 5018–5023. [Google Scholar] [CrossRef]
- Yang, C.; Li, Z.; Kang, W.; Tian, Y.; Yan, Y.; Chen, W. TET1 and TET3 are essential in induction of Th2-type immunity partly through regulation of IL-4/13A expression in zebrafish model. Gene 2016, 591, 201–208. [Google Scholar] [CrossRef]
- Collignon, E.; Canale, A.; Al Wardi, C.; Bizet, M.; Calonne, E.; Dedeurwaerder, S.; Garaud, S.; Naveaux, C.; Barham, W.; Wilson, A.; et al. Immunity drives TET1 regulation in cancer through NF-kappaB. Sci. Adv. 2018, 4, p7309. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Yuan, Z.; Dong, Z.; Peng, Y. Glutamine with probiotics attenuates intestinal inflammation and oxidative stress in a rat burn injury model through altered iNOS gene aberrant methylation. Am. J. Transl. Res. 2017, 9, 2535–2547. [Google Scholar] [PubMed]
- Liu, Z.; Gan, L.; Zhang, T.; Ren, Q.; Sun, C. Melatonin alleviates adipose inflammation through elevating alpha-ketoglutarate and diverting adipose-derived exosomes to macrophages in mice. J. Pineal Res. 2018, 64, e12455. [Google Scholar] [CrossRef]
- Shaknovich, R.; Cerchietti, L.; Tsikitas, L.; Kormaksson, M.; De, S.; Figueroa, M.E.; Ballon, G.; Yang, S.N.; Weinhold, N.; Reimers, M.; et al. DNA methyltransferase 1 and DNA methylation patterning contribute to germinal center B-cell differentiation. Blood 2011, 118, 3559–3569. [Google Scholar] [CrossRef]
- Unterberg, M.; Kreuzer, M.J.; Schafer, S.T.; Bazzi, Z.; Adamzik, M.; Rump, K. NFKB1 Promoter DNA from NT+402 to NT+99 Is Hypomethylated in Different Human Immune Cells. PLoS ONE 2016, 11, e156702. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.F.; Li, Q.P.; Dou, Y.X.; Wang, T.T.; Qu, C.; Liang, J.L.; Lin, Z.X.; Huang, X.Q.; Su, Z.R.; Chen, J.N.; et al. Therapeutic effect of Brucea javanica oil emulsion on experimental Crohn’s disease in rats: Involvement of TLR4/NF-kappaB signaling pathway. Biomed. Pharmacother. 2019, 114, 108766. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.W.; Lee, S.; Oh, B.M.; Lee, H.; Uhm, T.G.; Min, J.; Park, Y.; Yoon, S.R.; Kim, B.; Kim, J.W.; et al. Epigenetic modification of TLR4 promotes activation of NF-kappaB by regulating methyl-CpG-binding domain protein 2 and Sp1 in gastric cancer. Oncotarget 2016, 7, 4195–4209. [Google Scholar] [PubMed]
- Chanput, W.; Mes, J.J.; Savelkoul, H.F.; Wichers, H.J. Characterization of polarized THP-1 macrophages and polarizing ability of LPS and food compounds. Food Funct. 2013, 4, 266–276. [Google Scholar] [CrossRef] [PubMed]






| hTET1 siRNA | 5′–3′ Sense | 3′–5′ Antisense |
|---|---|---|
| #1 siRNA | CAGGAAGUUUCUGAUACCACCUCUU | AAGAGGUGGUAUCAGAAACUUCCUG |
| #2 siRNA | GGCUACACGAUUAGCUCCAAUUUAU | AUAAAUUGGAGCUAAUCGUGUAGCC |
| #3 siRNA | GGAAGCACUGUGGUUUGUACCUUAA | UUAAGGUACAAACCACAGUGCUUCC |
| Primer Name | 5′–3′ Forward | 5′–3′ Reverse |
|---|---|---|
| TET1 | GGCCCATATTATACACACCTTGG | GGACACCCATGAGAGCTTTTC |
| IL-6 | TGCAATAACCACCCCTGACC | AGCTGCGCAGAATGAGATGA |
| TNF-α | GCCTCTTCTCCTTCCTGATCG | TCGAGAAGATGATCTGACTGCC |
| CCL2 | AGAATCACCAGCAGCAAGTG | TCCATGGAATCCTGAACCCA |
| CCR7 | CATAGTCTTCCAGCTGCCCT | ACAAGAAAGGGTTGACGCAG |
| GAPDH | TCTCCTCTGACTTCAACAGCGACA | CCCTGTTGCTGTAGCCAAATTCGT |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.; Tian, C.; Li, Q.; Xu, Q. TET1 Knockdown Inhibits Porphyromonas gingivalis LPS/IFN-γ-Induced M1 Macrophage Polarization through the NF-κB Pathway in THP-1 Cells. Int. J. Mol. Sci. 2019, 20, 2023. https://doi.org/10.3390/ijms20082023
Huang Y, Tian C, Li Q, Xu Q. TET1 Knockdown Inhibits Porphyromonas gingivalis LPS/IFN-γ-Induced M1 Macrophage Polarization through the NF-κB Pathway in THP-1 Cells. International Journal of Molecular Sciences. 2019; 20(8):2023. https://doi.org/10.3390/ijms20082023
Chicago/Turabian StyleHuang, Yanlan, Cheng Tian, Qimeng Li, and Qiong Xu. 2019. "TET1 Knockdown Inhibits Porphyromonas gingivalis LPS/IFN-γ-Induced M1 Macrophage Polarization through the NF-κB Pathway in THP-1 Cells" International Journal of Molecular Sciences 20, no. 8: 2023. https://doi.org/10.3390/ijms20082023