Exploration of ABA Responsive miRNAs Reveals a New Hormone Signaling Crosstalk Pathway Regulating Root Growth of Populus euphratica
Abstract
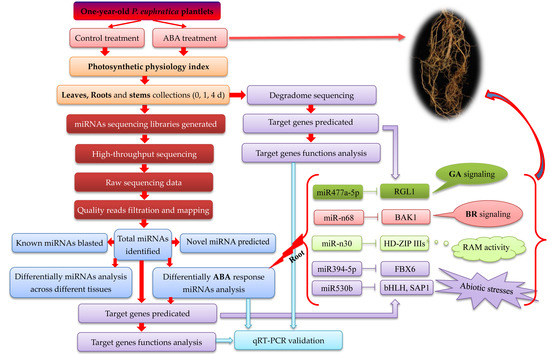
:1. Introduction
2. Results
2.1. Overview of Small RNAs (sRNAs) Responsive to ABA in the Roots and Stems of P. euphratica
2.2. Known miRNAs in P. euphratica Roots and Stems
2.3. Novel miRNAs in P. euphratica Roots and Stems
2.4. Differential miRNA Analysis between the Leaf, Root, and Stem of P. euphratica
2.5. Differentially Expressed miRNAs in Response to ABA in the Roots and Stems of P. euphratica
2.6. Validation of miRNAs by RT-qPCR
2.7. The Prediction and Validation of miRNA Target Genes in P. euphratica
3. Discussion
3.1. peu-miRNA477 Involved in the Crosstalk between ABA and GA in Root Growth
3.2. peu-miR-n68 Involved in the Crosstalk between ABA and BR in Root Growth
3.3. peu-miR-n30-Mediated Target Genes Involved in RAM Activity in Root Growth
3.4. peu-miR394-5p and peu-miR530b Mediated Stress-Related Genes Involved in Root Growth
4. Materials and Methods
4.1. Plant Materials and ABA Treatment
4.2. High-Throughput Sequencing of Small RNA
4.3. Sequencing Data Analysis for miRNA Identification and Annotation
4.4. Differential Expression Analysis of miRNAs Response to ABA in the Roots and Stems
4.5. RT-qPCR Validation
4.6. MiRNAs Targets Prediction by TargetFinder and Degradome Sequencing
4.7. Accession Number
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Lee, S.C.; Luan, S. ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Environ. 2012, 35, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef] [PubMed]
- Iuchi, S.; Kobayashi, M.; Taji, T.; Naramoto, M.; Seki, M.; Kato, T.; Tabata, S.; Kakubari, Y.; Yamaguchishinozaki, K.; Shinozaki, K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. Cell Mol. Biol. 2001, 27, 325–333. [Google Scholar] [CrossRef]
- Kushiro, T.; Okamoto, M.; Nakabayashi, K.; Yamagishi, K.; Kitamura, S.; Asami, T.; Hirai, N.; Koshiba, T.; Kamiya, Y.; Nambara, E. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: Key enzymes in ABA catabolism. EMBO J. 2014, 23, 1647–1656. [Google Scholar] [CrossRef] [PubMed]
- Christmann, A.; Moes, D.; Himmelbach, A.; Yang, Y.; Tang, Y.; Grill, E. Integration of abscisic acid signalling into plant responses. Plant Biol. 2006, 8, 314–325. [Google Scholar] [CrossRef] [PubMed]
- Raghavendra, A.S.; Gonugunta, V.K.; Christmann, A.; Grill, E. ABA perception and signalling. Trends Plant Sci. 2010, 15, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Yang, Z.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.F.F. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of start proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Guzman, M.; Pizzio, G.A.; Antoni, R.; Vera-Sirera, F.; Merilo, E.; Bassel, G.W.; Fernández, M.A.; Holdsworth, M.J.; Perez-Amador, M.A.; Kollist, H. Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 2012, 24, 2483–2496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkinson, S.; Davies, W.J. ABA-based chemical signalling: The co-ordination of responses to stress in plants. Plant Cell Environ. 2002, 25, 195–210. [Google Scholar] [CrossRef] [PubMed]
- Glinka, Z. Abscisic acid promotes both volume flow and ion release to the xylem in sunflower roots. Plant Physiol. 1980, 65, 537–540. [Google Scholar] [CrossRef] [PubMed]
- Sharp, R.E. Interaction with ethylene: Changing views on the role of abscisic acid in root and shoot growth responses to water stress. Plant Cell Environ. 2002, 25, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Sharp, R.E.; Lenoble, M.E. ABA, ethylene and the control of shoot and root growth under water stress. J. Exp. Bot. 2002, 53, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Achard, P.; Cheng, H.; de Grauwe, L.; Decat, J.; Schoutteten, H.; Moritz, T.; van der Straeten, D.; Peng, J.; Harberd, N.P. Integration of plant responses to environmentally activated phytohormonal signals. Science 2006, 311, 91–94. [Google Scholar] [CrossRef] [PubMed]
- Shu, K.; Zhou, W.; Yang, W. APETALA 2-domain-containing transcription factors: Focusing on abscisic acid and gibberellins antagonism. New Phytol. 2018, 217, 977–983. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Cai, Z.; Wang, X. The primary signaling outputs of brassinosteroids are regulated by abscisic acid signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 4543–4548. [Google Scholar] [CrossRef] [PubMed]
- Ha, Y.; Shang, Y.; Nam, K.H. Brassinosteroids modulate ABA-induced stomatal closure in Arabidopsis. J. Exp. Bot. 2016, 67, 6297–6308. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Li, C.; Wu, Z.; Jia, Y.; Wang, H.; Sun, S.; Mao, C.; Wang, X. Abscisic acid regulates auxin homeostasis in rice root tips to promote root hair elongation. Front. Plant Sci. 2017, 8, 1121. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Wang, P.; Wang, B.; Hsu, C.C.; Tang, K.; Zhang, H.; Hou, Y.J.; Zhao, Y.; Wang, Q.; Zhao, C. The SNRK2 kinases modulate miRNA accumulation in Arabidopsis. PLoS Genet. 2017, 13, e1006753. [Google Scholar] [CrossRef] [PubMed]
- Speth, C.; Willing, E.M.; Rausch, S.; Schneeberger, K.; Laubinger, S. RACK1 scaffold proteins influence miRNA abundance in Arabidopsis. Plant J. 2013, 76, 433–445. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Mou, W.; Luo, Z.; Li, L.; Jarukitt, L.; Mao, L.; Ying, T. Developmental and stress regulation on expression of a novel miRNA, Fan-miR73, and its target ABI5 in strawberry. Sci. Rep. 2016, 6, 23285. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.W.; Wang, L.J.; Mao, Y.B.; Cai, W.J.; Xue, H.W.; Chen, X.Y. Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell 2005, 17, 2204–2216. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.S.; Xie, Q.; Fei, J.F.; Chua, N.H. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 2005, 17, 1376–1386. [Google Scholar] [CrossRef] [PubMed]
- Robischon, M.; Du, J.; Miura, E.; Groover, A. The populus class III HD ZIP, popREVOLUTA, influences cambium initiation and patterning of woody stems. Plant Physiol. 2011, 155, 1214–1225. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Miura, E.; Robischon, M.; Martinez, C.; Groover, A. The populus class III HD ZIP transcription factor POPCORONA affects cell differentiation during secondary growth of woody stems. PLoS ONE 2011, 6, e17458. [Google Scholar] [CrossRef] [PubMed]
- Guddeti, S.; Zhang, D.C.; Li, A.L.; Leseberg, C.H.; Kang, H.; Li, X.G.; Zhai, W.X.; Johns, M.A.; Mao, L. Molecular evolution of the rice miR395 gene family. Cell Res. 2005, 15, 631–638. [Google Scholar] [CrossRef] [PubMed]
- Gang, L.; Yang, F.; Yu, D. MicroRNA395 mediates regulation of sulfate accumulation and allocation in Arabidopsis thaliana. Plant J. 2010, 62, 1046–1057. [Google Scholar]
- Sunkar, R.; Kapoor, A.; Zhu, J.K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Verma, S.; Trivedi, P.K. Involvement of small RNAs in phosphorus and sulfur sensing, signaling and stress: Current update. Front. Plant Sci. 2017, 8, 285. [Google Scholar] [CrossRef] [PubMed]
- Baek, D.; Kim, M.C.; Chun, H.J.; Kang, S.; Park, H.C.; Shin, G.; Park, J.; Shen, M.; Hong, H.; Kim, W.Y.; et al. Regulation of miR399f transcription by AtMYB2 affects phosphate starvation responses in Arabidopsis. Plant Physiol. 2013, 161, 362–373. [Google Scholar] [CrossRef] [PubMed]
- Luan, M.; Xu, M.; Lu, Y.; Zhang, Q.; Zhang, L.; Zhang, C.; Fan, Y.; Lang, Z.; Wang, L. Family-wide survey of miR169s and NF-YAs and their expression profiles response to abiotic stress in maize roots. PLoS ONE 2014, 9, e91369. [Google Scholar] [CrossRef] [PubMed]
- Jia, F.; Rock, C.D. miR846 and miR842 comprise a cistronic miRNA pair that is regulated by abscisic acid by alternative splicing in roots of Arabidopsis. Plant Mol. Biol. 2013, 81, 447–460. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.U.; Montgomery, T.A.; Fahlgren, N.; Kasschau, K.D.; Nonogaki, H.; Carrington, J.C. Repression of auxin response factor10 by microRNA160 is critical for seed germination and post-ermination stages. Plant J. Cell Mol. Biol. 2007, 52, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhao, C.; Li, J.; Liu, Z.; Wang, J. Root plasticity of Populus euphratica seedlings in response to different water table depths and contrasting sediment types. PLoS ONE 2015, 10, e0118691. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Q.; Ma, T.; Hu, Q.; Liu, B.; Wu, Y.; Zhou, H.; Wang, Q.; Wang, J.; Liu, J. Genome-scale transcriptome analysis of the desert poplar, Populus euphratica. Tree Physiol. 2011, 31, 452–461. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Qin, Y.; Hui, D.; Yin, W.; Xia, X. Genome-wide characterization of new and drought stress responsive microRNAs in Populus euphratica. J. Exp. Bot. 2011, 62, 3765. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Duan, H.; Li, J.; Deng, X.W.; Yin, W.; Xia, X. Global identification of miRNAs and targets in Populus euphratica under salt stress. Plant Mol. Biol. 2013, 81, 525–539. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, Y.; Ren, Y.; Xu, J.; Zhang, Z.; Wang, Y. Genome-wide identification of cold-responsive and new microRNAs in Populus tomentosa by high-throughput sequencing. Biochem. Biophys. Res. Commun. 2012, 417, 892–896. [Google Scholar] [CrossRef] [PubMed]
- Hui, D.; Xin, L.; Lian, C.; Yi, A.; Xia, X.; Yin, W. Genome-wide analysis of microRNA responses to the phytohormone abscisic acid in Populus euphratica. Front. Plant Sci. 2016, 7, 1184. [Google Scholar]
- Ma, T.; Wang, J.; Zhou, G.; Yue, Z.; Hu, Q.; Chen, Y.; Liu, B.; Qiu, Q.; Wang, Z.; Zhang, J. Genomic insights into salt adaptation in a desert poplar. Nat. Commun. 2013, 4, 2797. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, J.K. RNA-directed DNA methylation. Curr. Opin. Plant Biol. 2011, 14, 142–147. [Google Scholar] [CrossRef] [PubMed]
- He, X.J.; Ma, Z.Y.; Liu, Z.W. Non-coding RNA transcription and RNA-directed DNA methylation in Arabidopsis. Mol. Plant 2014, 7, 1406–1414. [Google Scholar] [CrossRef] [PubMed]
- Lewsey, M.G.; Hardcastle, T.J.; Melnyk, C.W.; Molnar, A.; Valli, A.; Urich, M.A.; Nery, J.R.; Baulcombe, D.C.; Ecker, J.R. Mobile small RNAs regulate genome-wide DNA methylation. Proc. Natl. Acad. Sci. USA 2016, 113, E801–E810. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Cui, Q.; Cao, Q.Z.; Liu, Q.; He, H.B.; Zhang, D.M.; Jia, G.X. Transcriptome-wide analysis of botrytis elliptica responsive microRNAs and their targets in lilium regale wilson by high-throughput sequencing and degradome analysis. Front. Plant Sci. 2017, 8, 753. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Zhu, J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019. [Google Scholar] [CrossRef] [PubMed]
- Lakhotia, N.; Joshi, G.; Bhardwaj, A.R.; Katiyar-Agarwal, S.; Agarwal, M.; Jagannath, A.; Goel, S.; Kumar, A. Identification and characterization of miRNAome in root, stem, leaf and tuber developmental stages of potato (Solanum tuberosum L.) by high-throughput sequencing. BMC Plant Biol. 2014, 14, 6. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, E.; Wuyts, J.; Rouzé, P.; Van de Peer, Y. Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics 2004, 20, 2911–2917. [Google Scholar] [CrossRef] [PubMed]
- Addo-Quaye, C.; Miller, W.A.; Michael, J. Cleaveland: A pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 2009, 25, 130–131. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.K.; Chang, C. Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 2002, 14, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Mhamdi, A.; Mauve, C.; Gouia, H.; Saindrenan, P.; Hodges, M.; Noctor, G. Cytosolic NADP-dependent isocitrate dehydrogenase contributes to redox homeostasis and the regulation of pathogen responses in Arabidopsis leaves. Plant Cell Environ. 2010, 33, 1112–1123. [Google Scholar] [PubMed]
- Lee, S.M.; Koh, H.J.; Park, D.C.; Song, B.J.; Huh, T.L.; Park, J.W. Cytosolic NADP(+)-dependent isocitrate dehydrogenase status modulates oxidative damage to cells. Free Radic. Biol. Med. 2002, 32, 1185–1196. [Google Scholar] [CrossRef]
- Fàbregas, N.; Li, N.; Boeren, S.; Nash, T.E.; Goshe, M.B.; Clouse, S.D.; de Vries, S.; Caño-Delgado, A.I. The brassinosteroid insensitive1-like3 signalosome complex regulates Arabidopsis root development. Plant Cell 2013, 25, 3377–3388. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Burd, S.; Lers, A. miR408 is involved in abiotic stress responses in Arabidopsis. Plant J. Cell Mol. Biol. 2015, 84, 169–187. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Wei, C.; Huang, J.; Shi, Y.; Wang, H.; Luo, Z.; Li, F.; Wang, R.; Yang, J.; Chen, J. Regulating expression of Cu/Zn-SOD gene in Nicotiana tabacum with miRNA398. Tob. Sci. Technol. 2014, 4, 99–102. [Google Scholar]
- Jin, D.; Wang, Y.; Zhao, Y.; Chen, M. MicroRNAs and their cross-talks in plant development. J. Genet. Genom. 2013, 40, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R. Abscisic acid synthesis and response. Arabidopsis Book 2013, 11, e0058. [Google Scholar] [CrossRef] [PubMed]
- Busov, V.; Meilan, R.; Pearce, D.W.; Rood, S.B.; Ma, C.; Tschaplinski, T.J.; Strauss, S.H. Transgenic modification of gai or rgl1 causes dwarfing and alters gibberellins, root growth, and metabolite profiles in Populus. Planta 2006, 224, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Van Esse, G.W.; van Mourik, S.; Stigter, H.; Colette, A.; Molenaar, J.; de Vries, S.C. A mathematical model for brassinosteroid insensitive1-mediated signaling in root growth and hypocotyl elongation. Plant Physiol. 2012, 160, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Gonzálezgarcía, M.P.; Vilarrasablasi, J.; Zhiponova, M.; Divol, F.; Moragarcía, S.; Russinova, E.; Cañodelgado, A.I. Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 2011, 138, 849–859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holton, N.; Caño-Delgado, A.; Harrison, K.; Montoya, T.; Chory, J.; Bishop, G.J. Tomato brassinosteroid insensitive1 is required for systemin-induced root elongation in Solanum pimpinellifolium but is not essential for wound signaling. Plant Cell 2007, 19, 1709–1717. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Singh, S.; Panigrahi, K.C.; Reski, R.; Sarkar, A.K. Balanced activity of microRNA166/165 and its target transcripts from the class III homeodomain-leucine zipper family regulates root growth in Arabidopsis thaliana. Plant Cell Rep. 2014, 33, 945–953. [Google Scholar] [CrossRef] [PubMed]
- Roberts, C.; Valdés, A.E.; Carlsbecker, A. Class III HD-ZIP transcription factors control root growth and vascular patterning. In Proceedings of the Plant Vascular Biology, Helsinki, Finland, 26–30 July 2013. [Google Scholar]
- Franco, J.A.; Bañón, S.; Vicente, M.J.; Miralles, J.; Martínezsánchez, J.J. Review article: Root development in horticultural plants grown under abiotic stress conditions—A review. J. Hortic. Sci. Biotechnol. 2011, 86, 543–556. [Google Scholar] [CrossRef]
- Khan, M.A.; Gemenet, D.C.; Villordon, A. Root system architecture and abiotic stress tolerance: Current knowledge in root and tuber crops. Front. Plant Sci. 2016, 7, 1584. [Google Scholar] [CrossRef] [PubMed]
- Litholdo, C.G.; Parker, B.L.; Eamens, A.L.; Larsen, M.R.; Cordwell, S.J.; Waterhouse, P.M. Proteomic identification of putative microRNA394 target genes in Arabidopsis thaliana identifies major latex protein family members critical for normal development. Mol. Cell. Proteom. 2016, 15, 2033–2047. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Nijhawan, A.; Arora, R.; Agarwal, P.; Ray, S.; Sharma, P.; Kapoor, S.; Tyagi, A.K.; Khurana, J.P. F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 2007, 143, 1467–1483. [Google Scholar] [CrossRef] [PubMed]
- Kohama, A.; Iida, K.; Oyamatsu, K. Overexpression of an f-box protein gene reduces abiotic stress tolerance and promotes root growth in rice. Mol. Plant 2011, 4, 190–197. [Google Scholar]
- Liu, W.; Tai, H.; Li, S.; Gao, W.; Zhao, M.; Xie, C.; Li, W.X. bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytol. 2014, 201, 1192–1204. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, H.; Jha, S.; Sharma, M.; Giri, J.; Tyagi, A.K. Rice SAPs are responsive to multiple biotic stresses and overexpression of OsSAP1, an A20/AN1 zinc-finger protein, enhances the basal resistance against pathogen infection in tobacco. Plant Sci. 2014, 225, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Roslan, N.F.; Abd Rashid, N.S.; Suka, I.E.; Nadiatul Ain, N.A.T.; Abdullah, N.S.; Asruri, M.B.; Toni, B.; Sukiran, N.L.; Zainal, Z.; Isa, N.M. Enhanced tolerance to salinity stress and ABA is regulated by Oryza sativa stress associated protein 8 (OsSAP 8). Aust. J. Crop Sci. 2017, 11, 853–860. [Google Scholar] [CrossRef]
- Zhou, Q.; Huang, Y.J.; Zeng, Y.R.; Mao, C.Z. Isolation of total RNA from embryo and endosperm of hickory nut and synthesis of cDNA. J. Zhejiang For. Sci. Technol. 2009, 29, 36–39. [Google Scholar]
- Jaakola, L.; Pirttilä, A.M.; Halonen, M.; Hohtola, A. Isolation of high quality RNA from bilberry (Vaccinium myrtillus L.) fruit. Mol. Biotechnol. 2001, 19, 201–203. [Google Scholar] [CrossRef]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. Mirdeepfinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Cui, Q.; Li, F.; Liu, A. Transcriptome-wide identification and characterization of microRNAs from castor bean (Ricinus communis L.). PLoS ONE 2013, 8, e69995. [Google Scholar] [CrossRef] [PubMed]
- Wen, M.; Shen, Y.; Shi, S.; Tang, T. miREvo: An integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinform. 2012, 13, 140. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, X. How to Use the DEGseq Package. Researchgate Net 2011. Available online: https://www.researchgate.net/publication/228762656_How_to_use_the_DEGseq_Package (accessed on 19 May 2018).
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate—A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar]
- Zhou, L.; Chen, J.; Li, Z.; Li, X.; Hu, X.; Huang, Y.; Zhao, X.; Liang, C.; Wang, Y.; Sun, L. Integrated profiling of microRNAs and mRNAs: MicroRNAs located on Xq27.3 associate with clear cell renal cell carcinoma. PLoS ONE 2010, 5, e15224. [Google Scholar] [CrossRef] [PubMed]
- Sturn, A.; Quackenbush, J.; Trajanoski, Z. Genesis: Cluster analysis of microarray data. Bioinformatics 2002, 18, 207–208. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Sun, Y.H.; Chiang, V.L. Stress-responsive microRNAs in Populus. Plant J. Cell Mol. Biol. 2008, 55, 131–151. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 2012, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Fahlgren, N. miRNA target prediction in plants. Methods Mol. Biol. 2010, 592, 51–57. [Google Scholar] [PubMed]
- Lavorgna, G.; Guffanti, A.; Borsani, G.; Ballabio, A.; Boncinelli, E. TargetFinder: Searching annotated sequence databases for target genes of transcription factors. Bioinformatics 1999, 15, 172–173. [Google Scholar] [CrossRef] [PubMed]
- German, M.A.; Luo, S.; Schroth, G.; Meyers, B.C.; Green, P.J. Construction of parallel analysis of RNA ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome. Nat. Protoc. 2009, 4, 356. [Google Scholar] [CrossRef] [PubMed]
- Addo-Quaye, C.; Miller, W.; Axtell, M.J. Cleaveland. Bioinformatics 2009, 25, 130–131. [Google Scholar] [CrossRef] [PubMed]










| Sample | Raw Data | Clean Reads | Length Filtered Reads | Mapped Reads | Length Filtered Unique Reads | Mapped Unique Reads |
|---|---|---|---|---|---|---|
| CS1 | 15,000,000 | 14,587,199 (97.25%) | 8,096,532 | 4,932,121 | 3,046,077 | 1,258,568 |
| CS2 | 12,295,483 | 11,416,579 (92.85%) | 5,998,393 | 2,391,734 | 3,216,544 | 855,550 |
| CS3 | 16,028,756 | 15,256,023 (95.18%) | 9,304,524 | 5,142,929 | 2,672,612 | 681,844 |
| SS1 | 12,314,579 | 12,051,738 (97.87%) | 4,489,652 | 2,836,870 | 1,742,854 | 722,719 |
| SS2 | 12,235,671 | 11,787,281 (96.34%) | 4,253,676 | 2,551,136 | 1,766,408 | 720,158 |
| SS3 | 10,721,576 | 10,189,259 (95.04%) | 3,307,671 | 1,850,502 | 1,428,834 | 539,275 |
| LS1 | 10,964,853 | 10,620,198 (96.86%) | 3,939,212 | 2,362,697 | 1,415,467 | 521,575 |
| LS2 | 11,332,951 | 10,519,899 (92.83%) | 1,440,580 | 659,872 | 580,173 | 156,926 |
| LS3 | 16,245,725 | 15,452,361 (95.12%) | 5,117,466 | 2,686,232 | 1,739,006 | 508,100 |
| CR1 | 17,920,873 | 16,698,033 (93.18%) | 9,605,828 | 1,555,617 | 3,616,174 | 334,181 |
| CR2 | 18,537,368 | 18,101,660 (97.65%) | 15,420,868 | 2,768,522 | 3,738,298 | 389,598 |
| CR3 | 11,075,762 | 10,302,814 (93.02%) | 5,578,796 | 1,395,324 | 2,345,761 | 347,958 |
| SR1 | 11,588,883 | 10,762,002 (92.86%) | 6,140,223 | 1,556,764 | 2,796,352 | 450,033 |
| SR2 | 10,966,078 | 10,312,261 (94.04%) | 8,254,705 | 2,095,221 | 3,544,129 | 538,110 |
| SR3 | 11,945,678 | 11,430,601 (95.69%) | 5,562,200 | 1,871,703 | 1,778,115 | 345,995 |
| LR1 | 12,971,014 | 12,601,308 (97.15%) | 6,650,163 | 3,399,255 | 2,706,102 | 777,214 |
| LR2 | 14,093,468 | 13,780,786 (97.78%) | 10,598,000 | 1,733,908 | 3,047,028 | 240,787 |
| LR3 | 10,769,268 | 10,361,201 (96.21%) | 5,914,549 | 1,497,009 | 1,906,663 | 234,996 |
| miRNA | Sequence (5′-3′) | LM (nt) | Reference miRNA | Family |
|---|---|---|---|---|
| peu-miR169l | AAGCCAAGGAUGACUUGCCUG | 21 | ptc-miR169o | |
| peu-miR169m | UAGCCAAGGAUGACUUGCUCG | 21 | ptc-miR169x | MIR169_1 |
| peu-miR171f | GGAUUGAGCCGCGCCAAUAUC | 21 | ptc-miR171k | MIR171_1 |
| peu-miR394-3p | CUGUUGGUCUCUCUUUGUAA | 20 | ptc-miR394a-5p | MIR394 |
| peu-miR399d | UGCCAAAGGAGAUUUGCCCCG | 21 | ptc-miR399a | MIR399 |
| peu-miR399e | UGCCAAAGAAGAUUUGCCCCG | 21 | ptc-miR399d | MIR399 |
| peu-miR399f | UGCCAAAGGAGAGUUGCCCUA | 21 | ptc-miR399i | MIR399 |
| peu-miR477c | GGAAACCUUUUGUGGGGGUUUG | 22 | ptc-miR477c | MIR477 |
| peu-miR6435 | UGAAUAAUGGAGACACUCUAG | 21 | ptc-miR6435 | |
| peu-miR6450 | CGAACACAGGACUCAAGGCUA | 21 | ptc-miR6450b | |
| peu-miR6472 | UAGUGAAUUCUAGGUCUCAAUC | 22 | ptc-miR6472 |
| miRNA | Sequences | miRNA* | Arm | LM(nt) | Location | Stand | MEF | LP(nt) | GC% | MEFI |
|---|---|---|---|---|---|---|---|---|---|---|
| miR-n95 | uuauuuaaauuugauuucuuu | No | 3p | 21 | scaffold35.1: 322060..322378 | + | −26.3 | 62 | 9.68% | 4.38 |
| miR-n96 | uuggaggaaauauauuuuggc | Yes | 3p | 21 | scaffold4.1: 1609767..1610085 | − | −38 | 84 | 38.10% | 1.19 |
| miR-n97 | ugaagagguagagaguguaauu | Yes | 5p | 22 | scaffold476.1: 74348..74667 | + | −67.7 | 146 | 47.26% | 0.98 |
| miR-n98 | gggacaaaaauggcauaagagg | No | 3p | 22 | scaffold98.1: 88683..89002 | − | −97.5 | 251 | 42.63% | 0.91 |
| miR-n99 | aaggaaaaugcauagaacaagu | No | 5p | 22 | scaffold32.1: 2021024..2021343 | + | −20 | 46 | 30.43% | 1.43 |
| miR-n100 | aauuuguacugugaaacu | No | 5p | 18 | scaffold462.1: 47666..47981 | + | −8.8 | 38 | 36.84% | 0.63 |
| miR-n101 | uauagaugacuauauuuagggagc | Yse | 5p | 24 | scaffold2579.1: 17957..18278 | − | −84.9 | 192 | 32.81% | 1.35 |
| Sample | Total Reads | Ratio | Unique Reads | Ratio |
|---|---|---|---|---|
| Raw Reads | 15,513,985 | / | 6,684,885 | / |
| Mappable Reads | 15,407,998 | 99.32% | 6,634,914 | 99.25% |
| Transcript Mapped Reads | 10,447,471 | 67.34% | 4,224,109 | 63.19% |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lian, C.; Yao, K.; Duan, H.; Li, Q.; Liu, C.; Yin, W.; Xia, X. Exploration of ABA Responsive miRNAs Reveals a New Hormone Signaling Crosstalk Pathway Regulating Root Growth of Populus euphratica. Int. J. Mol. Sci. 2018, 19, 1481. https://doi.org/10.3390/ijms19051481
Lian C, Yao K, Duan H, Li Q, Liu C, Yin W, Xia X. Exploration of ABA Responsive miRNAs Reveals a New Hormone Signaling Crosstalk Pathway Regulating Root Growth of Populus euphratica. International Journal of Molecular Sciences. 2018; 19(5):1481. https://doi.org/10.3390/ijms19051481
Chicago/Turabian StyleLian, Conglong, Kun Yao, Hui Duan, Qing Li, Chao Liu, Weilun Yin, and Xinli Xia. 2018. "Exploration of ABA Responsive miRNAs Reveals a New Hormone Signaling Crosstalk Pathway Regulating Root Growth of Populus euphratica" International Journal of Molecular Sciences 19, no. 5: 1481. https://doi.org/10.3390/ijms19051481