Genome-Wide Profiling of Small RNAs and Degradome Revealed Conserved Regulations of miRNAs on Auxin-Responsive Genes during Fruit Enlargement in Peaches
Abstract
:1. Introduction
2. Results
2.1. Genome-Wide Identification of miRNAs by Small RNA Sequencing in Peach
2.2. Experimental Verification of Important miRNAs in Peach
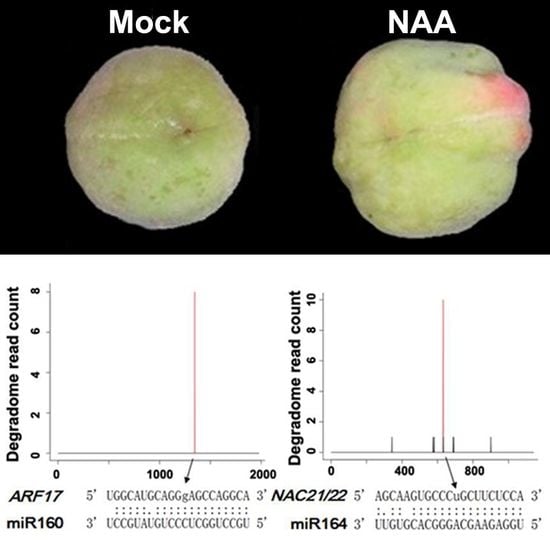
2.3. Genome-Wide Identification of miRNA Targets by Degradome Sequencing in Peach
2.4. Regulation of Crucial Genes by miRNAs During Fruit Development
3. Discussion
3.1. High-Throughput Sequencing Analysis
3.2. Identification of miRNAs in Peach Mesocarp
3.3. miRNA Targets in Auxin Signaling Pathways
3.4. miRNA Targets in Other Pathways
4. Materials and Methods
4.1. Plant Materials and NAA Treatment
4.2. Sample Embedding
4.3. Small RNA and Degradome RNA Library Construction
4.4. Bioinformatics Analysis for miRNA Identification
4.5. Stem-Loop RT-qPCR Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Verde, I.; Abbott, A.G.; Scalabrin, S.; Jung, S.; Shu, S.; Marroni, F.; Zhebentyayeva, T.; Dettori, M.T.; Grimwood, J.; Cattonaro, F. The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet. 2013, 45, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Verde, I.; Jenkins, J.; Dondini, L.; Micali, S.; Pagliarani, G.; Vendramin, E.; Paris, R.; Aramini, V.; Gazza, L.; Rossini, L. The Peach v2.0 release: High-resolution linkage mapping and deep resequencing improve chromosome-scale assembly and contiguity. BMC Genom. 2017, 18, 225. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Li, Y.-F.; Jagadeeswaran, G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012, 17, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. Plant microRNA: A small regulatory molecule with big impact. Dev. Biol. 2006, 289, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Curaba, J.; Singh, M.B.; Bhalla, P.L. miRNAs in the crosstalk between phytohormone signalling pathways. J. Exp. Bot. 2014, 65, 1425–1438. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B. MicroRNA: A new target for improving plant tolerance to abiotic stress. J. Exp. Bot. 2015, 66, 1749–1761. [Google Scholar] [CrossRef] [PubMed]
- Eldem, V.; Akçay, U.Ç.; Ozhuner, E.; Bakır, Y.; Uranbey, S.; Unver, T. Genome-wide identification of miRNAs responsive to drought in peach (Prunus persica) by high-throughput deep sequencing. PLoS ONE 2012, 7, e50298. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, M.; Yu, H.; Han, J.; Song, C.; Ma, R.; Fang, J. Computational identification of microRNAs in peach expressed sequence tags and validation of their precise sequences by miR-RACE. Mol. Biol. Rep. 2012, 39, 1975–1987. [Google Scholar] [CrossRef] [PubMed]
- Barakat, A.; Sriram, A.; Park, J.; Zhebentyayeva, T.; Main, D.; Abbott, A. Genome wide identification of chilling responsive microRNAs in Prunus persica. BMC Genom. 2012, 13, 481. [Google Scholar] [CrossRef] [PubMed]
- Solofoharivelo, M.; Walt, A.; Stephan, D.; Burger, J.; Murray, S. MicroRNAs in fruit trees: Discovery, diversity and future research directions. Plant Biol. 2014, 16, 856–865. [Google Scholar] [CrossRef] [PubMed]
- Vanneste, S.; Friml, J. Auxin: A trigger for change in plant development. Cell 2009, 136, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Li, S.B.; Xie, Z.Z.; Hu, C.G.; Zhang, J.Z. A Review of auxin response factors (ARFs) in plants. Front. Plant Sci. 2016, 7, 47. [Google Scholar] [CrossRef] [PubMed]
- Guilfoyle, T.J.; Hagen, G. Auxin response factors. Curr. Opin. Plant Biol. 2007, 10, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-W.; Wang, L.-J.; Mao, Y.-B.; Cai, W.-J.; Xue, H.-W.; Chen, X.-Y. Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell 2005, 17, 2204–2216. [Google Scholar] [CrossRef] [PubMed]
- Mallory, A.C.; Bartel, D.P.; Bartel, B. MicroRNA-directed regulation of Arabidopsis auxin response factor17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 2005, 17, 1360–1375. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, N.; Wang, H.; Kasahara, H.; Liu, J.; MacPherson, C.; Machida, Y.; Kamiya, Y.; Hannah, M.A.; Chua, N.-H. IAA-Ala Resistant3, an evolutionarily conserved target of miR167, mediates Arabidopsis root architecture changes during high osmotic stress. Plant Cell 2012, 24, 3590–3602. [Google Scholar] [CrossRef] [PubMed]
- Si-Ammour, A.; Windels, D.; Arn-Bouldoires, E.; Kutter, C.; Ailhas, J.; Meins, F., Jr.; Vazquez, F. miR393 and secondary siRNAs regulate expression of the TIR1/AFB2 auxin receptor clade and auxin-related development of Arabidopsis leaves. Plant Physiol. 2011, 157, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Marin, E.; Jouannet, V.; Herz, A.; Lokerse, A.S.; Weijers, D.; Vaucheret, H.; Nussaume, L.; Crespi, M.D.; Maizel, A. miR390, Arabidopsis TAS3 tasiRNAs, and their auxin response factor targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 2010, 22, 1104–1117. [Google Scholar] [CrossRef] [PubMed]
- Ripoll, J.J.; Bailey, L.J.; Mai, Q.-A.; Wu, S.L.; Hon, C.T.; Chapman, E.J.; Ditta, G.S.; Estelle, M.; Yanofsky, M.F. microRNA regulation of fruit growth. Nat. Plants 2015, 1, 15036. [Google Scholar] [CrossRef] [PubMed]
- Zlotorynski, E. Plant development: A fruit-bearing microRNA. Nat. Rev. Mol. Cell Biol. 2015, 16, 266. [Google Scholar] [CrossRef] [PubMed]
- Karlova, R.; van Haarst, J.C.; Maliepaard, C.; van de Geest, H.; Bovy, A.G.; Lammers, M.; Angenent, G.C.; de Maagd, R.A. Identification of microRNA targets in tomato fruit development using high-throughput sequencing and degradome analysis. J. Exp. Bot. 2013, 64, 1863–1878. [Google Scholar] [CrossRef] [PubMed]
- Navarro, L.; Dunoyer, P.; Jay, F.; Arnold, B.; Dharmasiri, N.; Estelle, M.; Voinnet, O.; Jones, J.D. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 2006, 312, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Agusti, M.; Almela, V.; Andreu, I.; Juan, M.; Zacarias, L. Synthetic auxin 3,5,6-TPA promotes fruit development and climacteric in Prunus persica L. Batsch. J. Hortic. Sci. Biotechnol. 1999, 74, 556–560. [Google Scholar] [CrossRef]
- Stern, R.A.; Flaishman, M.; Ben-Arie, R. Effect of synthetic auxins on fruit size of five cultivars of Japanese plum (Prunus salicina Lindl.). Sci. Hortic. 2007, 112, 304–309. [Google Scholar] [CrossRef]
- Agusti, M.; Gariglio, N.; Castillo, A.; Juan, M.; Almela, V.; Martinez-Fuentes, A.; Mesejo, C. Effect of the synthetic auxin 2,4-DP on fruit development of loquat. Plant Growth Regul. 2003, 41, 129–132. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, X.; Liu, J.; Kiba, T.; Woo, J.; Ojo, T.; Hafner, M.; Tuschl, T.; Chua, N.H.; Wang, X.J. Deep sequencing of small RNAs specifically associated with Arabidopsis AGO1 and AGO4 uncovers new AGO functions. Plant J. 2011, 67, 292–304. [Google Scholar] [CrossRef] [PubMed]
- Denman, R.B. Using RNAFOLD to predict the activity of small catalytic RNAs. Biotechniques 1993, 15, 1090–1095. [Google Scholar] [PubMed]
- Meyers, B.C.; Axtell, M.J.; Bartel, B.; Bartel, D.P.; Baulcombe, D.; Bowman, J.L.; Cao, X.; Carrington, J.C.; Chen, X.; Green, P.J.; et al. Criteria for annotation of plant MicroRNAs. Plant Cell 2008, 20, 3186–3190. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Chu, C. MicroRNAs in crop improvement: Fine-tuners for complex traits. Nat. Plants 2017, 3, 17077. [Google Scholar] [CrossRef] [PubMed]
- Samad, A.F.A.; Sajad, M.; Nazaruddin, N.; Fauzi, I.A.; Murad, A.M.A.; Zainal, Z.; Ismail, I. MicroRNA and Transcription Factor: Key Players in Plant Regulatory Network. Front. Plant Sci. 2017, 8, 565. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Addo-Quaye, C.; Eshoo, T.W.; Bartel, D.P.; Axtell, M.J. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 2008, 18, 758–762. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 2011, 39, W155–W159. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.P.; Montgomery, T.A.; Fahlgren, N.; Kasschau, K.D.; Nonogaki, H.; Carrington, J.C. Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 2007, 52, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, F.T.; Timmermans, M.C. An interplay between small regulatory RNAs patterns leaves. Plant Signal. Behav. 2007, 2, 519–521. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.-X.; Zhang, L.-F.; Li, W.-F.; Qi, L.-W.; Han, S.-Y. MIR166a Affects the Germination of Somatic Embryos in Larixleptolepis by Modulating IAA Biosynthesis and Signaling Genes. J. Plant Growth Regul. 2017, 36, 1–8. [Google Scholar] [CrossRef]
- Guo, H.-S.; Xie, Q.; Fei, J.-F.; Chua, N.-H. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 2005, 17, 1376–1386. [Google Scholar] [CrossRef] [PubMed]
- Jones-Rhoades, M.W.; Bartel, D.P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 2004, 14, 787–799. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Zhu, J.-K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.-F.; Tian, Q.; Reed, J.W. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 2006, 133, 4211–4218. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.H.; Han, S.J.; Yoon, E.K.; Lee, W.S. Evidence of an auxin signal pathway, microRNA167-ARF8-GH3, and its response to exogenous auxin in cultured rice cells. Nucleic Acids Res. 2006, 34, 1892–1899. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Potuschak, T.; Colón-Carmona, A.; Gutiérrez, R.A.; Doerner, P. Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc. Natl. Acad. Sci. USA 2005, 102, 12978–12983. [Google Scholar] [CrossRef] [PubMed]
- Guan, Q.; Lu, X.; Zeng, H.; Zhang, Y.; Zhu, J. Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J. 2013, 74, 840–851. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Ding, Y.; Liu, H. MiR398 and plant stress responses. Physiol. Plant. 2011, 143, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Jagadeeswaran, G.; Saini, A.; Sunkar, R. Biotic and abiotic stress down-regulate miR398 expression in Arabidopsis. Planta 2009, 229, 1009–1014. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Wang, W.-X.; Ren, L.; Chen, Q.-J.; Mendu, V.; Willcut, B.; Dinkins, R.; Tang, X.; Tang, G. Differential and dynamic regulation of miR398 in response to ABA and salt stress in Populustremula and Arabidopsis thaliana. Plant Mol. Biol. 2009, 71, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.; Zhao, M.; Gao, W.; Sun, S.; Li, W.X. microRNA/microRNA* complementarity is important for the regulation pattern of NFYA5 by miR169 under dehydration shock in Arabidopsis. Plant J. 2017, 91, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Manning, K.; Tor, M.; Poole, M.; Hong, Y.; Thompson, A.J.; King, G.J.; Giovannoni, J.J.; Seymour, G.B. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 2006, 38, 948–952. [Google Scholar] [CrossRef] [PubMed]
- Moxon, S.; Jing, R.; Szittya, G.; Schwach, F.; Rusholme Pilcher, R.L.; Moulton, V.; Dalmay, T. Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res. 2008, 18, 1602–1609. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zou, Z.; Zhang, J.; Zhang, Y.; Han, Q.; Hu, T.; Xu, X.; Liu, H.; Li, H.; Ye, Z. Over-expression of sly-miR156a in tomato results in multiple vegetative and reproductive trait alterations and partial phenocopy of the sft mutant. FEBS Lett. 2011, 585, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Li, Z.; Miao, Q.; Yang, Y.; Deng, W.; Hao, Y. The auxin receptor homologue in Solanum lycopersicum stimulates tomato fruit set and leaf morphogenesis. J. Exp. Bot. 2011, 62, 2815–2826. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef] [PubMed]
- Giannoulatou, E.; Park, S.H.; Humphreys, D.T.; Ho, J.W. Verification and validation of bioinformatics software without a gold standard: A case study of BWA and Bowtie. BMC Bioinform. 2014, 15, S15. [Google Scholar] [CrossRef] [PubMed]
- Addo-Quaye, C.; Miller, W.; Axtell, M.J. CleaveLand: A pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 2009, 25, 130–131. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R. Real-time quantification of microRNAs by stem–loop RT–PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef] [PubMed]
- Varkonyi-Gasic, E.; Wu, R.; Wood, M.; Walton, E.F.; Hellens, R.P. Protocol: A highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 2007, 3, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]



| miRNA ID | Target ID | Mock Cleavage Site | Mock Confidence | Mock p-Value | Mock Count | NAA Cleavage Site | NAA Confidence | NAA p-Value | NAA Count | Target Annotation |
|---|---|---|---|---|---|---|---|---|---|---|
| miR156 | ppa012607m | 791 | High | 8.00 × 10-3 | 5 | N/A 1 | N/A | N/A | N/A | Squamosa promoter-binding protein 2 |
| miR156 | ppa009498m | 63 | Intemediate | 2.82 × 10-2 | 0.3 | N/A | N/A | N/A | N/A | Homeobox-leucine zipper protein ATHB-13 |
| miR156 | ppa013510m | 629 | Intemediate | 4.42 × 10-2 | 0.5 | N/A | N/A | N/A | N/A | Auxin-repressed 12.5 kDa protein |
| miR156 | ppa010764m | N/A | N/A | N/A | N/A | 19 | high | 2.02 × 10-2 | 3 | Inhibitor of growth protein 5 |
| miR157 | ppa012607m | 791 | High | 5.54 × 10-3 | 5 | N/A | N/A | N/A | N/A | Squamosa promoter-binding protein 2 |
| miR160 | ppa003136m | 1346 | High | 2.09 × 10-3 | 8 | 1346 | high | 3.00 × 10-3 | 3 | Auxin response factor 17 |
| miR164 | ppa008801m | 632 | High | 1.36 × 10-3 | 8 | 632 | high | 1.83 × 10-3 | 10 | NAC domain-containing protein 21/22 |
| miR165/miR166 | ppa001378m | 565 | High | 3.70 × 10-3 | 2 | N/A | N/A | N/A | N/A | Homeobox-leucine zipper protein REVOLUTA |
| miR165/miR166 | ppa001386m | 574 | High | 3.70 × 10 -3 | 6.5 | N/A | N/A | N/A | N/A | Homeobox-leucine zipper protein ATHB-8 |
| miR165/miR166 | ppa001343m | 589 | High | 3.70 × 10-3 | 38 | N/A | N/A | N/A | N/A | Homeobox-leucine zipper protein ATHB-14 |
| miR165/miR166 | ppa001405m | 615 | High | 9.88 × 10-3 | 6.5 | N/A | N/A | N/A | N/A | Homeobox-leucine zipper protein ATHB-15 |
| miR168 | ppa000547m | N/A | N/A | N/A | N/A | 478 | high | 2.50 × 10-3 | 4 | Protein argonaute 1B |
| miR169 | ppa006634m | 1252 | High | 1.73 × 10-3 | 8 | N/A | N/A | N/A | N/A | Nuclear transcription factor Y subunit A-1 |
| miR169 | ppa012173m | N/A | N/A | N/A | N/A | 709 | high | 1.83 × 10-3 | 2 | Nuclear transcription factor Y subunit A-7 |
| miR169 | ppa008065m | N/A | N/A | N/A | N/A | 1069 | high | 4.03 × 10-3 | 2 | Protein TIFY 6B |
| miR172 | ppa021782m | N/A | N/A | N/A | N/A | 1234 | high | 1.83 × 10-3 | 2 | Ethylene-responsive transcription factor RAP2-7 |
| miR393 | ppa003465m | 1612 | High | 1.85 × 10-3 | 6 | 1612 | high | 2.50 × 10-3 | 11 | Protein AUXIN SIGNALING F-BOX 2 |
| miR394 | ppa004699m | 2100 | Intemediate | 1.47 × 10-2 | 2 | N/A | N/A | N/A | N/A | F-box only protein 6 |
| miR396 | ppa021277m | 452 | High | 1.28 × 10-3 | 2 | 452 | high | 2.46 × 10-3 | 2 | AtGRF9 (GROWTH-REGULATING FACTOR 9) |
| miR396 | ppa019623m | 749 | High | 1.28 × 10-2 | 2 | N/A | N/A | N/A | N/A | AtGRF1 (GROWTH-REGULATING FACTOR 1) |
| miR396 | ppa019752m | N/A | N/A | N/A | N/A | 716 | high | 2.30 × 10-2 | 3 | AtGRF8 (GROWTH-REGULATING FACTOR 8) |
| miR405b-p3 | ppa008493m | 71 | High | 4.15 × 10-2 | 2 | N/A | N/A | N/A | N/A | Myb-like protein J |
| miR482-p5 | ppa000031m | N/A | N/A | N/A | N/A | 4660 | high | 2.94 × 10-2 | 2 | Chromodomain-helicase-DNA-binding protein 5 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, M.; Hu, X.; Wei, Y.; Hou, X.; Yuan, X.; Liu, J.; Liu, Y. Genome-Wide Profiling of Small RNAs and Degradome Revealed Conserved Regulations of miRNAs on Auxin-Responsive Genes during Fruit Enlargement in Peaches. Int. J. Mol. Sci. 2017, 18, 2599. https://doi.org/10.3390/ijms18122599
Shi M, Hu X, Wei Y, Hou X, Yuan X, Liu J, Liu Y. Genome-Wide Profiling of Small RNAs and Degradome Revealed Conserved Regulations of miRNAs on Auxin-Responsive Genes during Fruit Enlargement in Peaches. International Journal of Molecular Sciences. 2017; 18(12):2599. https://doi.org/10.3390/ijms18122599
Chicago/Turabian StyleShi, Mengya, Xiao Hu, Yu Wei, Xu Hou, Xue Yuan, Jun Liu, and Yueping Liu. 2017. "Genome-Wide Profiling of Small RNAs and Degradome Revealed Conserved Regulations of miRNAs on Auxin-Responsive Genes during Fruit Enlargement in Peaches" International Journal of Molecular Sciences 18, no. 12: 2599. https://doi.org/10.3390/ijms18122599