Kinases Involved in Both Autophagy and Mitosis
Abstract
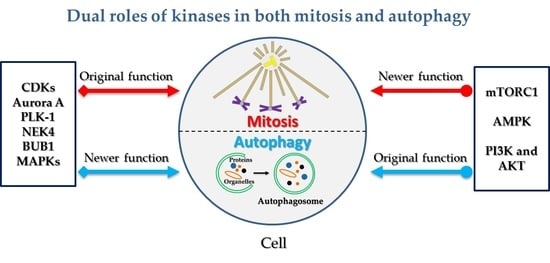
:1. Introduction
2. Kinases Originally Involved in Mitotic Regulation Are Shown to Play Additional Roles in Autophagy
2.1. Cyclin-Dependent Kinases
2.2. Aurora Kinases
2.3. Polo-Like Kinases
2.4. NimA-Related Protein Kinase 4 (NEK4 Kinase)
2.5. Budding Uninhibited by Benzimidazoles 1 (BUB1) Kinase
2.6. Mitogen-Activated Protein Kinases (MAPKs)
3. Kinases Originally Involved in Autophagy Regulation Are Shown to Play Additional Roles in Mitosis
3.1. Mechanistic Target of Rapamycin Complex 1 (mTORC1)
3.2. AMP Activated Protein Kinase (AMPK)
3.3. Phosphoinositide-3 Kinase (PI3K) and Protein Kinase B (AKT)
4. Conclusions
5. Perspectives
Acknowledgments
Conflicts of Interest
References
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Acevedo, A.A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2016, 12, 1–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, H.; Osawa, T.; Fujioka, Y.; Noda, N.N. Structural biology of the core autophagy machinery. Curr. Opin. Struct. Biol. 2017, 43, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ji, X.; Wang, D.; Liu, J.; Zhang, X. Autophagic flux is highly active in early mitosis and differentially regulated throughout the cell cycle. Oncotarget 2016, 7, 39705–39718. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, X. Autophagy in mitotic animal cells. Sci. Bull. 2016, 61, 105–107. [Google Scholar] [CrossRef]
- Brown, N.R.; Korolchuk, S.; Martin, M.P.; Stanley, W.A.; Moukhametzianov, R.; Noble, M.E.; Endicott, J.A. CDK1 structures reveal conserved and unique features of the essential cell cycle CDK. Nat. Commun. 2015, 6, 6769. [Google Scholar] [CrossRef] [PubMed]
- Nigg, E.A. The substrates of the CDC2 kinase. Semin. Cell Biol. 1991, 2, 261–270. [Google Scholar] [PubMed]
- Kõivomägi, M.; Valk, E.; Venta, R.; Iofik, A.; Lepiku, M.; Morgan, D.O.; Loog, M. Dynamics of Cdk1 Substrate Specificity during the Cell Cycle. Mol. Cell 2011, 42, 610–623. [Google Scholar] [CrossRef] [PubMed]
- Mathiassen, S.G.; De Zio, D.; Cecconi, F. Autophagy and the cell cycle: A complex landscape. Fron. Oncol. 2017, 7, 51. [Google Scholar] [CrossRef] [PubMed]
- Sridharan, S.; Jain, K.; Basu, A. Regulation of autophagy by kinases. Cancers 2011, 3, 2630–2654. [Google Scholar] [CrossRef] [PubMed]
- Malumbres, M. Cyclin-dependent kinases. Genome Biol. 2014, 15, 122. [Google Scholar] [CrossRef] [PubMed]
- Linares, J.F.; Amanchy, R.; Diaz-Meco, M.T.; Moscat, J. Phosphorylation of p62 by cdk1 controls the timely transit of cells through mitosis and tumor cell proliferation. Mol. Cell Biol. 2011, 31, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Furuya, T.; Kim, M.; Lipinski, M.; Li, J.; Kim, D.; Lu, T.; Shen, Y.; Rameh, L.; Yankner, B.; Tsai, L.H.; et al. Negative regulation of Vps34 by Cdk mediated phosphorylation. Mol. Cell 2010, 38, 500–511. [Google Scholar] [CrossRef] [PubMed]
- Rubinsztein, D.C. Cdks regulate autophagy via Vps34. Mol. Cell 2010, 38, 483–484. [Google Scholar] [CrossRef] [PubMed]
- Bunnell, B.A.; Heath, L.S.; Adams, D.E.; Lahti, J.M.; Kidd, V.J. Increased expression of a 58-kDa protein kinase leads to changes in the CHO cell cycle. Proc. Natl. Acad. Sci. USA 1990, 87, 7467–7471. [Google Scholar] [CrossRef] [PubMed]
- Trembley, J.H.; Loyer, P.; Hu, D.; Li, T.; Grenet, J.; Lahti, J.M.; Kidd, V.J. Cyclin dependent kinase 11 in RNA transcription and splicing. Prog. Nucleic Acid Res. Mol. Biol. 2003, 77, 263–288. [Google Scholar]
- Cornelis, S.; Bruynooghe, Y.; Denecker, G.; Van Huffel, S.; Tinton, S.; Beyaert, R. Identification and characterization of a novel cell cycle–regulated internal ribosome entry site. Mol. Cell 2000, 5, 597–605. [Google Scholar] [CrossRef]
- Hu, D.; Valentine, M.; Kidd, V.J.; Lahti, J.M. CDK11(p58) is required for the maintenance of sister chromatid cohesion. J. Cell Sci. 2007, 120, 2424–2434. [Google Scholar] [CrossRef] [PubMed]
- Petretti, C.; Savoian, M.; Montembault, E.; Glover, D.M.; Prigent, C.; Giet, R. The PITSLRE/CDK11 p58 protein kinase promotes centrosome maturation and bipolar spindle formation. EMBO Rep. 2006, 7, 418–424. [Google Scholar] [PubMed]
- Wilker, E.W.; van Vugt, M.A.; Artim, S.A.; Huang, P.H.; Petersen, C.P.; Reinhardt, H.C.; Feng, Y.; Sharp, P.A.; Sonenberg, N.; White, F.M.; et al. 14-3-3 δ controls mitotic translation to facilitate cytokinesis. Nature 2007, 446, 329–332. [Google Scholar] [CrossRef] [PubMed]
- Franck, N.; Montembault, E.; Rome, P.; Pascal, A.; Cremet, J.-Y.; Giet, R. CDK11p58 is required for centriole duplication and Plk4 recruitment to mitotic centrosomes. PLoS ONE 2011, 6, e14600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rakkaa, T.; Escudé, C.; Giet, R.; Magnaghi, -J.L.; Jaulin, C. CDK11p58 kinase activity is required to protect sister chromatid cohesion at centromeres in mitosis. Chromosome Res. 2014, 22, 267–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Inoue, A.; Lahti, J.M.; Kidd, V.J. Failure to proliferate and mitotic arrest of CDK11p110/p58-null mutant mice at the blastocyst stage of embryonic cell development. Mol. Cell. Biol. 2004, 24, 3188–3197. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, S.; Croft, D.R.; O’Prey, J.; Meedendorp, A.; O’Prey, M.; Dufès, C.; Ryan, K.M. The cyclin-dependent kinase PITSLRE/CDK11 is required for successful autophagy. Autophagy 2011, 7, 1295–1301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Shen, J.K.; Hornicek, F.J.; Kan, Q.; Duan, Z. The emerging roles and therapeutic potential of cyclin-dependent kinase 11 (CDK11) in human cancer. Oncotarget 2016, 7, 40846–40859. [Google Scholar] [CrossRef] [PubMed]
- Chandramouli, A.; Shi, J.; Feng, Y.; Holubec, H.; Shanas, R.M.; Bhattacharyya, A.K.; Zheng, W.; Nelson, M.A. Haploinsufficiency of the cdc2l gene contributes to skin cancer development in mice. Carcinogenesis 2007, 28, 2028–2035. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.; Huang, S.; Wang, L.; Zhou, R.; Wang, L.; Xiao, X.; Li, D.; Cai, Y.; Zhou, X.; Wu, J. CDK11p58 inhibits ERα-positive breast cancer invasion by targeting integrin β3 via the repression of ERα signaling. BMC Cancer 2014, 14, 577. [Google Scholar] [CrossRef] [PubMed]
- Duan, Z.; Zhang, J.; Choy, E.; Harmon, D.; Liu, X.; Nielsen, P.; Mankin, H.; Gray, N.S.; Hornicek, F.J. Systematic kinome shRNA screening identifies CDK11 (PITSLRE) kinase expression is critical for osteosarcoma cell growth and proliferation. Clin. Cancer Res. 2012, 18, 4580–4588. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Choy, E.; Cote, G.; Harmon, D.; Ye, S.; Kan, Q.; Mankin, H.; Hornicek, F.; Duan, Z. Cyclin-dependent kinase 11 (CDK11) is crucial in the growth of liposarcoma cells. Cancer Lett. 2014, 342, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.; Wang, L.; Xiao, X.; Wei, P.; Wang, Y.; Zhou, X. Abnormal expression of CDK11 p58 in prostate cancer. Cancer Cell Int. 2014, 14, 2. [Google Scholar] [CrossRef] [PubMed]
- Haq, R.; Randall, S.; Midmer, M.; Yee, K.; Zanke, B. NKIATRE is a novel conserved cdc2-related kinase. Genomics 2001, 71, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Yee, K.W.; Moore, S.J.; Midmer, M.; Zanke, B.W.; Tong, F.; Hedley, D.; Minden, M.D. NKIAMRE, a novel conserved CDC2-related kinase with features of both mitogen-activated protein kinases and cyclin-dependent kinases. Biochem. Biophys. Res. Commun. 2003, 308, 784–792. [Google Scholar] [CrossRef]
- Szyniarowski, P.; Corcelle-Termeau, E.; Farkas, T.; Høyer, H.M.; Nylandsted, J.; Kallunki, T.; Jäättelä, M. A comprehensive siRNA screen for kinases that suppress macroautophagy in optimal growth conditions. Autophagy 2011, 7, 892–903. [Google Scholar] [CrossRef] [PubMed]
- Capparelli, C.; Chiavarina, B.; Whitaker-Menezes, D.; Pestell, T.G.; Pestell, R.G.; Hulit, J.; Andò, S.; Howell, A.; Martinez-Outschoorn, U.E.; Sotgia, F. CDK inhibitors (p16/p19/p21) induce senescence and autophagy in cancer-associated fibroblasts,“fueling” tumor growth via paracrine interactions, without an increase in neo-angiogenesis. Cell Cycle 2012, 11, 3599–3610. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, K.; Daido, S.; Yamamoto, A.; Kobayashi, R.; Yokoyama, T.; Aoki, H.; Iwado, E.; Shinojima, N.; Kondo, Y.; Kondo, S. Pivotal role of the cyclin-dependent kinase inhibitor p21WAF1/CIP1 in apoptosis and autophagy. J. Biol. Chem. 2008, 283, 388–397. [Google Scholar] [CrossRef] [PubMed]
- Krenn, V.; Musacchio, A. The Aurora B kinase in chromosome Bi-orientation and spindle checkpoint signaling. Front. Oncol. 2015, 5, 225. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Bian, M.; Jiang, Q.; Zhang, C. Roles of Aurora kinases in mitosis and tumorigenesis. Mol. Cancer Res. 2007, 5, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Su, C.-L.; Fan, Y.-W. Induction of autophagy and inhibition of oncoprotein Aurora-A activity in curcumin-enhanced chemosensitivity of Grade III Human Bladder Cancer T24 cells to FDA-approved anticancer drugs. FASEB J. 2015, 29, 752.1. [Google Scholar]
- Xu, L.Z.; Long, Z.J.; Peng, F.; Liu, Y.; Xu, J.; Wang, C.; Jiang, L.; Guo, T.; Kamran, M.; Li, S.S. Aurora kinase a suppresses metabolic stress-induced autophagic cell death by activating mTOR signaling in breast cancer cells. Oncotarget 2014, 5, 7498–7511. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Li, H.; Yan, X.G.; Zhou, Z.W.; Yi, Z.G.; He, Z.X.; Pan, S.T.; Yang, Y.X.; Wang, Z.Z.; Zhang, X.; et al. Alisertib induces cell cycle arrest and autophagy and suppresses epithelial-to-mesenchymal transition involving PI3K/Akt/mTOR and sirtuin 1-mediated signaling pathways in human pancreatic cancer cells. Drug Des. Devel. Ther. 2015, 9, 575–601. [Google Scholar] [PubMed]
- Liu, Z.; Wang, F.; Zhou, Z.W.; Xia, H.C.; Wang, X.Y.; Yang, Y.X.; He, Z.X.; Sun, T.; Zhou, S.F. Alisertib induces G2/M arrest, apoptosis, and autophagy via PI3K/Akt/mTOR-and p38 MAPK-mediated pathways in human glioblastoma cells. Am. J. Transl. Res. 2017, 9, 845–873. [Google Scholar] [PubMed]
- Ding, Y.h.; Zhou, Z.W.; Ha, C.F.; Zhang, X.Y.; Pan, S.T.; He, Z.X.; Edelman, J.L.; Wang, D.; Yang, Y.X.; Zhang, X.; et al. Alisertib, an Aurora kinase A inhibitor, induces apoptosis and autophagy but inhibits epithelial to mesenchymal transition in human epithelial ovarian cancer cells. Drug Des. Devel. Ther. 2015, 9, 425–464. [Google Scholar] [PubMed]
- Zhu, Q.; Yu, X.; Zhou, Z.W.; Zhou, C.; Chen, X.W.; Zhou, S.F. Inhibition of aurora a kinase by alisertib induces autophagy and cell cycle arrest and increases chemosensitivity in human hepatocellular carcinoma HepG2 cells. Curr. Cancer Drug Targets 2017, 17, 386–401. [Google Scholar] [CrossRef] [PubMed]
- Boss, D.S.; Beijnen, J.H.; Schellens, J.H. Clinical experience with aurora kinase inhibitors: A review. Oncologist 2009, 14, 780–793. [Google Scholar] [CrossRef] [PubMed]
- Archambault, V.; Lepine, G.; Kachaner, D. Understanding the polo kinase machine. Oncogene 2015, 34, 4799–4807. [Google Scholar] [CrossRef] [PubMed]
- Lens, S.M.; Voest, E.E.; Medema, R.H. Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nature reviews. Cancer 2010, 10, 825–841. [Google Scholar] [PubMed]
- Ruf, S.; Heberle, A.M.; Langelaar, -M.M.; Gelino, S.; Wilkinson, D.; Gerbeth, C.; Schwarz, J.J.; Holzwarth, B.; Warscheid, B.; Meisinger, C.; et al. PLK1 (polo like kinase 1) inhibits MTOR complex 1 and promotes autophagy. Autophagy 2017, 13, 486–505. [Google Scholar] [CrossRef] [PubMed]
- Valianou, M.; Cox, A.M.; Pichette, B.; Hartley, S.; Paladhi, U.R.; Astrinidis, A. Pharmacological inhibition of Polo-like kinase 1 (PLK1) by BI-2536 decreases the viability and survival of hamartin and tuberin deficient cells via induction of apoptosis and attenuation of autophagy. Cell Cycle 2015, 14, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Deeraksa, A.; Pan, J.; Sha, Y.; Liu, X.D.; Eissa, N.T.; Lin, S.H.; Yu-Lee, L.Y. Plk1 is upregulated in androgen-insensitive prostate cancer cells and its inhibition leads to necroptosis. Oncogene 2013, 32, 2973–2983. [Google Scholar] [CrossRef] [PubMed]
- Nigg, E.A. Mitotic kinases as regulators of cell division and its checkpoints. Nature reviews. Mol. Cell Biol. 2001, 2, 21–32. [Google Scholar]
- Nguyen, C.L.; Possemato, R.; Bauerlein, E.L.; Xie, A.; Scully, R.; Hahn, W.C. Nek4 regulates entry into replicative senescence and the response to DNA damage in human fibroblasts. Mol. Cell. Biol. 2012, 32, 3963–3977. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Jo, D.S.; Jo, S.Y.; Shin, D.W.; Shim, S.; Jo, Y.K.; Shin, J.H.; Ha, Y.J.; Jeong, S.Y.; Hwang, J.J.; et al. Inhibition of never in mitosis A (NIMA)-related kinase-4 reduces survivin expression and sensitizes cancer cells to TRAIL-induced cell death. Oncotarget 2016, 7, 65957–65967. [Google Scholar] [PubMed]
- Bolanos-Garcia, V.M.; Kiyomitsu, T.; D'Arcy, S.; Chirgadze, D.Y.; Grossmann, J.G.; Matak-Vinkovic, D.; Venkitaraman, A.R.; Yanagida, M.; Robinson, C.V.; Blundell, T.L. The crystal structure of the N-terminal region of BUB1 provides insight into the mechanism of BUB1 recruitment to kinetochores. Structure 2009, 17, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Qi, W.; Yu, H. KEN-box-dependent degradation of the Bub1 spindle checkpoint kinase by the anaphase-promoting complex/cyclosome. J. Biol. Chem. 2007, 282, 3672–3679. [Google Scholar] [CrossRef] [PubMed]
- Bolanos-Garcia, V.M.; Blundell, T.L. BUB1 and BUBR1: Multifaceted kinases of the cell cycle. Trends Biochem. Sci. 2011, 36, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Niikura, Y.; Dixit, A.; Scott, R.; Perkins, G.; Kitagawa, K. BUB1 mediation of caspase-independent mitotic death determines cell fate. J. Cell Biol. 2007, 178, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Santaguida, S.; Vasile, E.; White, E.; Amon, A. Aneuploidy-induced cellular stresses limit autophagic degradation. Genes Dev. 2015, 29, 2010–2021. [Google Scholar] [CrossRef] [PubMed]
- Kyriakis, J.M.; Avruch, J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev. 2001, 81, 807–869. [Google Scholar] [PubMed]
- Chen, Z.; Gibson, T.B.; Robinson, F.; Silvestro, L.; Pearson, G.; Xu, B.; Wright, A.; Vanderbilt, C.; Cobb, M.H. MAP kinases. Chem. Rev. 2001, 101, 2449–2476. [Google Scholar] [CrossRef] [PubMed]
- Pearson, G.; Robinson, F.; Gibson, T.B.; Xu, B.; Karandikar, M.; Berman, K.; Cobb, M.H. Mitogen-activated protein (MAP) kinase pathways: Regulation and physiological functions. Endocr. Rev. 2001, 22, 153–183. [Google Scholar] [CrossRef] [PubMed]
- Sui, X.; Kong, N.; Ye, L.; Han, W.; Zhou, J.; Zhang, Q.; He, C.; Pan, H. p38 and JNK MAPK pathways control the balance of apoptosis and autophagy in response to chemotherapeutic agents. Cancer Lett. 2014, 344, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Cha, H.; Wang, X.; Li, H.; Fornace, A.J. A functional role for p38 MAPK in modulating mitotic transit in the absence of stress. J. Biol. Chem. 2007, 282, 22984–22992. [Google Scholar] [CrossRef] [PubMed]
- Bulavin, D.V.; Amundson, S.A.; Fornace, A.J. p38 and Chk1 kinases: Different conductors for the G2/M checkpoint symphony. Curr. Opin. Genet. Dev. 2002, 12, 92–97. [Google Scholar] [CrossRef]
- Mikhailov, A.; Shinohara, M.; Rieder, C.L. The p38-mediated stress-activated checkpoint: A rapid response system for delaying progression through antephase and entry into mitosis. Cell Cycle 2005, 4, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Kukkonen, M.A.; Sicora, O.; Kaczynska, K.; Oetken, L.C.; Pouwels, J.; Laine, L.; Kallio, M.J. Loss of p38γ MAPK induces pleiotropic mitotic defects and massive cell death. J. Cell Sci. 2011, 124, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Cui, Q.; Tashiro, S.; Onodera, S.; Minami, M.; Ikejima, T. Oridonin induced autophagy in human cervical carcinoma HeLa cells through Ras, JNK, and P38 regulation. J. Pharmacol. Sci. 2007, 105, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Zou, M.J.; Zhao, L.; Lu, N.; Sun, Y.J.; Gou, S.H.; Xi, T.; Guo, Q.L. E Platinum, a newly synthesized platinum compound, induces autophagy via inhibiting phosphorylation of mTOR in gastric carcinoma BGC-823 cells. Toxicol. Lett. 2012, 210, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Zhou, J.; Sheng, F.; Zhu, H.; Deng, X.; Xia, B.; Lin, J. The heme oxygenase-1 inhibitor ZnPPIX induces non-canonical, Beclin 1-independent, autophagy through p38 MAPK pathway. Acta Biochim. Biophys. Sin. 2012, 44, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.C.; Yu, L.; Wang, H.J.; Tashiro, S.I.; Onodera, S.; Ikejima, T. TNFα-induced necroptosis and autophagy via supression of the p38–NF-κB survival pathway in L929 cells. J. Pharmacol. Sci. 2011, 117, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Thyagarajan, A.; Jedinak, A.; Nguyen, H.; Terry, C.; Baldridge, L.A.; Jiang, J.; Sliva, D. Triterpenes from Ganoderma Lucidum induce autophagy in colon cancer through the inhibition of p38 mitogen-activated kinase (p38 MAPK). Nutr. Cancer 2010, 62, 630–640. [Google Scholar] [CrossRef] [PubMed]
- Keil, E.; Höcker, R.; Schuster, M.; Essmann, F.; Ueffing, N.; Hoffman, B.; Liebermann, D.; Pfeffer, K.; Schulze, O.K.; Schmitz, I. Phosphorylation of Atg5 by the Gadd45β–MEKK4-p38 pathway inhibits autophagy. Cell Death Differ. 2013, 20, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Webber, J.L.; Tooze, S.A. Coordinated regulation of autophagy by p38α MAPK through mAtg9 and p38IP. EMBO J. 2010, 29, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.H.; Lee, B.H.; Ahn, S.G.; Oh, S.H. Proteasome inhibition-induced p38 MAPK/ERK signaling regulates autophagy and apoptosis through the dual phosphorylation of glycogen synthase kinase 3β. Biochem. Biophys. Res. Commun. 2012, 418, 759–764. [Google Scholar] [CrossRef] [PubMed]
- Chambard, J.C.; Lefloch, R.; Pouysségur, J.; Lenormand, P. ERK implication in cell cycle regulation. Biochim. Biophys. Acta 2007, 1773, 1299–1310. [Google Scholar] [CrossRef] [PubMed]
- Tang, N.; Marshall, W.F.; McMahon, M.; Metzger, R.J.; Martin, G.R. Control of mitotic spindle angle by the RAS-regulated ERK1/2 pathway determines lung tube shape. Science 2011, 333, 342–345. [Google Scholar] [CrossRef] [PubMed]
- Bravo-San Pedro, J.M.; Gómez, S.R.; Niso, S.M.; Pizarro, E.E.; Aiastui, P.A.; Gorostidi, A.; Climent, V.; Lopez de Maturana, R.; Sanchez, P.R.; Lopez de Munain, A.; et al. The MAPK1/3 pathway is essential for the deregulation of autophagy observed in G2019S LRRK2 mutant fibroblasts. Autophagy 2012, 8, 1537–1539. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Liu, H.; Yu, J.; Zhao, Z.; Xiao, F.; Xia, T.; Wang, C.; Li, K.; Deng, J.; Guo, Y.; et al. MAPK1/3 regulate hepatic lipid metabolism via ATG7-dependent autophagy. Autophagy 2016, 12, 592–593. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, X.; Ma, K.; Yang, J.; Zhou, J.; Fu, W.; Wei, F.; Wang, L.; Zhu, W.G. The axis of MAPK1/3-XBP1u-FOXO1 controls autophagic dynamics in cancer cells. Autophagy 2013, 9, 794–796. [Google Scholar] [CrossRef] [PubMed]
- Houel, -R.L.; Philippe, L.; Piquemal, M.; Ciapa, B. Autophagy is used as a survival program in unfertilized sea urchin eggs that are destined to die by apoptosis after inactivation of MAPK1/3 (ERK2/1). Autophagy 2013, 9, 1527–1539. [Google Scholar] [CrossRef] [PubMed]
- Oktay, K.; Buyuk, E.; Oktem, O.; Oktay, M.; Giancotti, F.G. The c-Jun N-terminal kinase JNK functions upstream of Aurora B to promote entry into mitosis. Cell Cycle 2008, 7, 533–541. [Google Scholar] [CrossRef] [PubMed]
- Almuedo, -C.M.; Crespo, X.; Seebeck, F.; Bartscherer, K.; Salò, E.; Adell, T. JNK controls the onset of mitosis in planarian stem cells and triggers apoptotic cell death required for regeneration and remodeling. PLoS Genet. 2014, 10, e1004400. [Google Scholar]
- Bogoyevitch, M.A.; Yeap, Y.Y.; Qu, Z.; Ngoei, K.R.; Yip, Y.Y.; Zhao, T.T.; Heng, J.I.; Ng, D.C. WD40-repeat protein 62 is a JNK-phosphorylated spindle pole protein required for spindle maintenance and timely mitotic progression. J. Cell Sci. 2012, 125, 5096–5109. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, G.J.; Tsuji, T.; Chen, M.; Jiang, W.; Ronai, Z.A. Interplay between Cdh1 and JNK activity during the cell cycle. Nat. Cell Biol. 2010, 12, 686–695. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, G.J.; Tsuji, T.; Cross, J.V.; Davis, R.J.; Templeton, D.J.; Jiang, W.; Ze’ev, A.R. JNK-mediated phosphorylation of Cdc25C regulates cell cycle entry and G2/M DNA damage checkpoint. J. Biol. Chem. 2010, 285, 14217–14228. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Yang, M.; Yang, L.; Yu, Y.; Xie, M.; Zhu, S.; Kang, R.; Tang, D.; Jiang, Z.; Yuan, W.; et al. HMGB1 regulates autophagy through increasing transcriptional activities of JNK and ERK in human myeloid leukemia cells. BMB Rep. 2011, 44, 601–606. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Shu, W.; Dai, W.; Gao, B.; Xiong, S. ROS-mediated JNK activation contributes to HBx-induced autophagy via regulating Beclin-1/Bcl-2 interaction. J. Virol. 2017, 91, JVI.00001-17. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Li, D.; Wang, L.; Xia, L.; Ma, J.; Guan, Z.; Feng, G.; Zhu, X. c-Jun NH2-terminal kinase activation is essential for up-regulation of LC3 during ceramide-induced autophagy in human nasopharyngeal carcinoma cells. J. Transl. Med. 2011, 9, 161. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Johnson, D.E. Bortezomib induces autophagy in head and neck squamous cell carcinoma cells via JNK activation. Cancer Lett. 2012, 314, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Yang, Y.; Xing, D. Bcl-2 and Bcl-xL play important roles in the crosstalk between autophagy and apoptosis. FEBS J. 2011, 278, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Garcia, -A.M.; Zhao, R.; Puri, C.; Toh, P.P.; Sadiq, O.; Rubinsztein, D.C. Bim inhibits autophagy by recruiting Beclin 1 to microtubules. Mol. Cell 2012, 47, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Sinha, S.C.; Levine, B. Dual role of JNK1-mediated phosphorylation of Bcl-2 in autophagy and apoptosis regulation. Autophagy 2008, 4, 949–951. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.H.; Noh, J.H.; Kim, J.K.; Eun, J.W.; Bae, H.J.; Chang, Y.G.; Kim, M.G.; Park, W.S.; Lee, J.Y.; Lee, S.Y.; et al. Histone deacetylase 6 functions as a tumor suppressor by activating c-Jun NH2-terminal kinase-mediated beclin 1-dependent autophagic cell death in liver cancer. Hepatology 2012, 56, 644–657. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, L.; Deng, R.; Tang, J.; Shen, Y.; Guo, J.; Wang, Y.; Xia, L.; Feng, G.; Liu, Q.; et al. The pivotal role of c-Jun NH2-terminal kinase-mediated Beclin 1 expression during anticancer agents-induced autophagy in cancer cells. Oncogene 2009, 28, 886–898. [Google Scholar] [CrossRef] [PubMed]
- Lorin, S.; Pierron, G.; Ryan, K.M.; Codogno, P.; Djavaheri, M.M. Evidence for the interplay between JNK and p53-DRAM signaling pathways in the regulation of autophagy. Autophagy 2010, 6, 153–154. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; Wu, X.Q.; Deng, R.; Sun, T.; Feng, G.K.; Zhu, X.F. Upregulation of sestrin 2 expression via JNK pathway activation contributes to autophagy induction in cancer cells. Cell Signal. 2013, 25, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Singh, R.; Xiang, Y.; Czaja, M.J. Macroautophagy and chaperone-mediated autophagy are required for hepatocyte resistance to oxidant stress. Hepatology 2010, 52, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Abe, M.K.; Saelzler, M.P.; Espinosa, R.; Kahle, K.T.; Hershenson, M.B.; Le Beau, M.M.; Rosner, M.R. ERK8, a New Member of the Mitogen-activated Protein Kinase Family. J. Biol. Chem. 2002, 277, 16733–16743. [Google Scholar] [CrossRef] [PubMed]
- Colecchia, D.; Strambi, A.; Sanzone, S.; Iavarone, C.; Rossi, M.; Dall’Armi, C.; Piccioni, F.; Verrotti di, P.; Chiariello, M. MAPK15/ERK8 stimulates autophagy by interacting with LC3 and GABARAP proteins. Autophagy 2012, 8, 1724–1740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laplante, M.; Sabatini, D.M. mTOR signaling at a glance. J. Cell. Sci. 2009, 122, 3589–3594. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.H.; Ro, S.H.; Cao, J.; Otto, N.M.; Kim, D.H. mTOR regulation of autophagy. FEBS Lett. 2010, 584, 1287–1295. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Puente, C.; Hendrickson, R.C.; Jiang, X. Nutrient-regulated phosphorylation of ATG13 inhibits starvation-induced autophagy. J. Biol. Chem. 2016, 291, 6026–6035. [Google Scholar] [CrossRef] [PubMed]
- Lopez, B.E.; Vazquez, M.A.; Pérez-Martínez, M.C.; Oliveras, F.C.; Pérez, B.F.; Bernadó, L.; Menendez, J.A. Serine 2481-autophosphorylation of mammalian target of rapamycin (mTOR) couples with chromosome condensation and segregation during mitosis: Confocal microscopy characterization and immunohistochemical validation of PP-mTORSer2481 as a novel high-contrast mitosis marker in breast cancer core biopsies. Int. J. Oncol. 2010, 36, 107–115. [Google Scholar]
- Vazquez, M.A.; Sauri, N.T.; Menendez, O.J.; Oliveras, F.C.; Cufí, S.; Corominas, F.B.; López, B.E.; Menendez, J.A. Ser2481-autophosphorylated mTOR colocalizes with chromosomal passenger proteins during mammalian cell cytokinesis. Cell Cycle 2012, 11, 4211–4221. [Google Scholar] [CrossRef] [PubMed]
- Kogasaka, Y.; Hoshino, Y.; Hiradate, Y.; Tanemura, K.; Sato, E. Distribution and association of mTOR with its cofactors, raptor and rictor, in cumulus cells and oocytes during meiotic maturation in mice. Mol. Reprod. Dev. 2013, 80, 334–348. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, -V.F.; Badura, M.L.; Braunstein, S.; Narasimhan, M.; Schneider, R.J. Mitotic raptor promotes mTORC1 activity, G2/M cell cycle progression, and internal ribosome entry site-mediated mRNA translation. Mol. Cell. Biol. 2010, 30, 3151–3164. [Google Scholar]
- Gwinn, D.M.; Asara, J.M.; Shaw, R.J. Raptor is phosphorylated by cdc2 during mitosis. PLoS ONE 2010, 5, e9197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shuda, M.; Chang, Y.; Moore, P.S. Mitotic 4E-BP1 hyperphosphorylation and cap-dependent translation. Cell Cycle 2015, 14, 3005–3006. [Google Scholar] [CrossRef] [PubMed]
- Mihaylova, M.M.; Shaw, R.J. The AMPK signaling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 2011, 13, 1016–1023. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Ji, J.; Yan, X.H. Cross-talk between AMPK and mTOR in regulating energy balance. Crit. Rev. Food Sci. Nutr. 2012, 52, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Egan, D.F.; Shackelford, D.B.; Mihaylova, M.M.; Gelino, S.; Kohnz, R.A.; Mair, W.; Vasquez, D.S.; Joshi, A.; Gwinn, D.M.; Taylor, R.; et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 2011, 331, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Shang, L.; Chen, S.; Du, F.; Li, S.; Zhao, L.; Wang, X. Nutrient starvation elicits an acute autophagic response mediated by Ulk1 dephosphorylation and its subsequent dissociation from AMPK. Proc. Natl. Acad. Sci. USA 2011, 108, 4788–4793. [Google Scholar] [CrossRef] [PubMed]
- Roach, P.J. AMPK→ULK1→Autophagy. Mol. Cell. Biol. 2011, 31, 3082–3084. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, -M.A.; López, -B.E.; Oliveras, -F.C.; Pérez-Martínez, M.C.; Bernadó, L.; Menendez, J.A. Mitotic kinase dynamics of the active form of AMPK (phospho-AMPKαThr172) in human cancer cells. Cell Cycle 2009, 8, 788–791. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, -M.A.; Oliveras, -F.C.; Menendez, J.A. The active form of the metabolic sensor AMP-activated protein kinase α (AMPKα) directly binds the mitotic apparatus and travels from centrosomes to the spindle midzone during mitosis and cytokinesis. Cell Cycle 2009, 8, 2385–2398. [Google Scholar] [CrossRef] [PubMed]
- Thaiparambil, J.T.; Eggers, C.M.; Marcus, A.I. AMPK regulates mitotic spindle orientation through phosphorylation of myosin regulatory light chain. Mol. Cell. Biol. 2012, 32, 3203–3217. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, M.A.; Oliveras, F.C.; Cufí, S.; Menendez, J.A. Polo-like kinase 1 regulates activation of AMP-activated protein kinase (AMPK) at the mitotic apparatus. Cell Cycle 2011, 10, 1295–1302. [Google Scholar] [CrossRef] [PubMed]
- Banko, M.R.; Allen, J.J.; Schaffer, B.E.; Wilker, E.W.; Tsou, P.; White, J.L.; Villén, J.; Wang, B.; Kim, S.R.; Sakamoto, K.; et al. Chemical genetic screen for AMPKα2 substrates uncovers a network of proteins involved in mitosis. Mol. Cell 2011, 44, 878–892. [Google Scholar] [CrossRef] [PubMed]
- Cantley, L.C. The phosphoinositide 3-kinase pathway. Science 2002, 296, 1655–1657. [Google Scholar] [CrossRef] [PubMed]
- Stokoe, D.; Stephens, L.R.; Copeland, T.; Gaffney, P.R.; Reese, C.B.; Painter, G.F.; Holmes, A.B.; McCormick, F.; Hawkins, P.T. Dual role of phosphatidylinositol-3, 4, 5-trisphosphate in the activation of protein kinase B. Science 1997, 277, 567–570. [Google Scholar] [CrossRef] [PubMed]
- Sarbassov, D.D.; Guertin, D.A.; Ali, S.M.; Sabatini, D.M. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 2005, 307, 1098–1101. [Google Scholar] [CrossRef] [PubMed]
- Ronan, B.; Flamand, O.; Vescovi, L.; Dureuil, C.; Durand, L.; Fassy, F.; Bachelot, M.-F.; Lamberton, A.; Mathieu, M.; Bertrand, T.; et al. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat. Chem. Biol. 2014, 10, 1013–1019. [Google Scholar] [CrossRef] [PubMed]
- Jaber, N.; Dou, Z.; Chen, J.S.; Catanzaro, J.; Jiang, Y.P.; Ballou, L.M.; Selinger, E.; Ouyang, X.; Lin, R.Z.; Zhang, J.; et al. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc. Natl. Acad. Sci. USA 2012, 109, 2003–2008. [Google Scholar] [CrossRef] [PubMed]
- Shanware, N.P.; Bray, K.; Abraham, R.T. The PI3K, metabolic, and autophagy networks: Interactive partners in cellular health and disease. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 89–106. [Google Scholar] [CrossRef] [PubMed]
- Roberts, E.C.; Shapiro, P.S.; Nahreini, T.S.; Pages, G.; Pouyssegur, J.; Ahn, N.G. Distinct cell cycle timing requirements for extracellular signal-regulated kinase and phosphoinositide 3-kinase signaling pathways in somatic cell mitosis. Mol. Cell. Biol. 2002, 22, 7226–7241. [Google Scholar] [CrossRef] [PubMed]
- Shtivelman, E.; Sussman, J.; Stokoe, D. A role for PI 3-kinase and PKB activity in the G2/M phase of the cell cycle. Curr. Biol. 2002, 12, 919–924. [Google Scholar] [CrossRef]
- Ornelas, I.M.; Silva, T.M.; Fragel, M.L.; Ventura, A.L.M. Inhibition of PI3K/Akt pathway impairs G2/M transition of cell cycle in late developing progenitors of the avian embryo retina. PLoS ONE 2013, 8, e53517. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Shi, Y.; Woods, K.W.; Hessler, P.; Kroeger, P.; Wilsbacher, J.; Wang, J.; Wang, J.Y.; Li, C.; Li, Q.; et al. Akt inhibitor a-443654 interferes with mitotic progression by regulating aurora a kinase expression. Neoplasia 2008, 10, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Campa, C.C.; Martini, M.; De Santis, M.C.; Hirsch, E. How PI3K-derived lipids control cell division. Front. Cell Dev. Biol. 2015, 3, 61. [Google Scholar] [CrossRef] [PubMed]
- Sagona, A.P.; Nezis, I.P.; Pedersen, N.M.; Liestøl, K.; Poulton, J.; Rusten, T.E.; Skotheim, R.I.; Raiborg, C.; Stenmark, H. PtdIns (3) P controls cytokinesis through KIF13A-mediated recruitment of FYVE-CENT to the midbody. Nat. Cell Biol. 2010, 12, 362–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thoresen, S.B.; Pedersen, N.M.; Liestøl, K.; Stenmark, H. A phosphatidylinositol 3-kinase class III sub-complex containing VPS15, VPS34, Beclin 1, UVRAG and BIF-1 regulates cytokinesis and degradative endocytic traffic. Exp. Cell Res. 2010, 316, 3368–3378. [Google Scholar] [CrossRef] [PubMed]
- Janetopoulos, C.; Borleis, J.; Vazquez, F.; Iijima, M.; Devreotes, P. Temporal and spatial regulation of phosphoinositide signaling mediates cytokinesis. Dev. Cell 2005, 8, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Nezis, I.P.; Sagona, A.P.; Schink, K.O.; Stenmark, H. Divide and ProsPer: The emerging role of PtdIns3P in cytokinesis. Trends Cell Biol. 2010, 20, 642–649. [Google Scholar] [CrossRef] [PubMed]
- Boularan, C.; Kamenyeva, O.; Cho, H.; Kehrl, J.H. Resistance to inhibitors of cholinesterase (Ric)-8A and Gαi contribute to cytokinesis abscission by controlling vacuolar protein-sorting (Vps) 34 activity. PLoS ONE 2014, 9, e86680. [Google Scholar] [CrossRef] [PubMed]
- Lamb, C.A.; Yoshimori, T.; Tooze, S.A. The autophagosome: Origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 2013, 14, 759–774. [Google Scholar] [CrossRef] [PubMed]
- Caballe, A.; Wenzel, D.M.; Agromayor, M.; Alam, S.L.; Skalicky, J.J.; Kloc, M.; Carlton, J.G.; Labrador, L.; Sundquist, W.I.; Martin, S.J. ULK3 regulates cytokinetic abscission by phosphorylating ESCRT-III proteins. Elife 2015, 4, e06547. [Google Scholar] [CrossRef] [PubMed]
- Young, A.R.; Narita, M.; Ferreira, M.; Kirschner, K.; Sadaie, M.; Darot, J.F.; Tavaré, S.; Arakawa, S.; Shimizu, S.; Watt, F.M. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009, 23, 798–803. [Google Scholar] [CrossRef] [PubMed]
| Kinases | Originally Found to Function in Mitosis or Autophagy | Later Evidences for Their Function in the Other Process |
|---|---|---|
| Aurora A | Mitotic spindle formation and centrosome separation [35,36] | Autophagy inhibition [37,38,39,40,41,42] |
| CDKs | Cell cycle regulation (primarily in mitosis by CDK1 and CDK11) [5,22] | Autophagy regulation [12,13,23] |
| PLK1 | G2/M transition [45,46] | Autophagy induction [46,47]. |
| NEK-4 | Cell cycle arrest [50] | Autophagy inhibition [32] |
| BUB1 | Spindle assembly checkpoint and chromosome alignment [53,54] | Autophagy inhibition [32,55] |
| MAPKs | P38: mitotic entry [64], Erk1/2: G2/M transition [73,74], JNK: onset of mitosis and mitotic spindle regulation [79,80,81] | P38: dual roles in autophagy regulation [65,66,67,68,69], Erk1/2: autophagy regulation [75,76,77,78], JNK: autophagy induction [84,85,86,87] |
| mTORC1 | Starvation induced autophagy [100,101] | Mitotic progression [102,103,104,105,106,107] |
| AMPK | Energy associated autophagy [110,111,112] | Mitotic spindle orientation [113,114,115,116,117] |
| PI3K/AKT | Induce or inhibit autophagy [119,120,121,122,123] | G2/Mitosis transition and cytokinesis [124,125,126,127,128,129,130,131,132] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Zhang, X. Kinases Involved in Both Autophagy and Mitosis. Int. J. Mol. Sci. 2017, 18, 1884. https://doi.org/10.3390/ijms18091884
Li Z, Zhang X. Kinases Involved in Both Autophagy and Mitosis. International Journal of Molecular Sciences. 2017; 18(9):1884. https://doi.org/10.3390/ijms18091884
Chicago/Turabian StyleLi, Zhiyuan, and Xin Zhang. 2017. "Kinases Involved in Both Autophagy and Mitosis" International Journal of Molecular Sciences 18, no. 9: 1884. https://doi.org/10.3390/ijms18091884