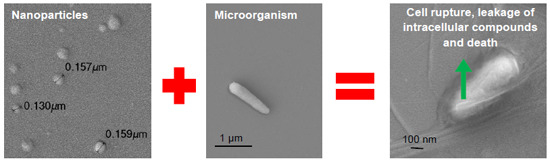
Supramolecular Cationic Assemblies against Multidrug-Resistant Microorganisms: Activity and Mechanism of Action
Abstract
:1. Introduction
2. Results and Discussion
| Dispersion | [DODAB] (mM) | [CMC] (mg/mL) | [PDDA] (mg/mL) | Dz (nm) | ζ (mV) | P |
|---|---|---|---|---|---|---|
| DODAB BF | 1.0 | - | - | 81 ± 1 | 52 ± 3 | 0.172 ± 0.010 |
| DODAB BF/CMC | 0.1 | 0.1 | - | 114 ± 1 | −43 ± 2 | 0.125 ± 0.010 |
| DODAB BF/CMC/PDDA | 0.1 | 0.1 | 0.1 | 108 ± 1 | 37 ± 2 | 0.131 ± 0.010 |


| MDR Microorganism | [DODAB] (μM) | [PDDA] (μg/mL) | [PDDA]in NP (μg/mL) |
|---|---|---|---|
| P. aeruginosa MDR | 10 | 1.5 | 2.5 * |
| K. pneumoniae KPC+ | >1000 | 0.9 | 2.5 * |
| MRSA | >1000 | 5.0 | 8.0 ** |
| C. albicans fluconazole R | >1000 | 0.8 | 2.5 * |
| Microorganism | Number of Cells | Absorbance | Nanomoles Pi |
|---|---|---|---|
| P. aeruginosa MDR | 2.1 × 107 | 0.052 ± 0.003 | 5.2 ± 0.3 |
| 1.6 × 109 | 0.441 ± 0.010 | 44.1 ± 1.0 | |
| K. pneumoniae KPC+ | 8.6 × 106 | 0.045 ± 0.010 | 4.5 ± 1.0 |
| 1.2 × 108 | 0.216 ± 0.006 | 21.6 ± 0.6 | |
| MRSA | 9.1 × 106 | 0.032 ± 0.004 | 3.2 ± 0.4 |
| 6.3 × 1010 | 0.260 ± 0.001 | 26.0 ± 0.1 | |
| C. albicans fluconazole R | 8.9 × 104 | 0.008 ± 0.002 | 0.8 ± 0.2 |
| 3.3 × 106 | 0.401 ± 0.010 | 40.1 ± 1.0 |


3. Experimental Section
3.1. Materials
3.2. Preparation and Characterization of the Hybrid Cationic Nanoparticles
3.3. Growth of Multidrug Resistant and Reference (ATCC) Microorganisms
3.4. Determination of Cell Viability
3.5. Determination of Leakage of Phosphorylated Compounds from the Cells
3.6. Visualization of NP, Cells and Cells/NP by Scanning Electron Microscopy
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Mitragotri, S.; Burke, P.A.; Langer, R. Overcoming the challenges in administering biopharmaceuticals: Formulation and delivery strategies. Nat. Rev. Drug Discov. 2014, 13, 655–672. [Google Scholar] [CrossRef] [PubMed]
- Chaubey, P.; Patel, R.R.; Mishra, B. Development and optimization of curcumin-loaded mannosylated chitosan nanoparticles using response surface methodology in the treatment of visceral leishmaniasis. Expert Opin. Drug Deliv. 2014, 11, 1163–1181. [Google Scholar] [CrossRef] [PubMed]
- Mura, S.; Zouhiri, F.; Lerondel, S.; Maksimenko, A.; Mougin, J.; Gueutin, C.; Brambilla, D.; Caron, J.; Sliwinski, E.; Lepape, A.; et al. Novel isoprenoyl nanoassembled prodrug for Paclitaxel delivery. Bioconjug. Chem. 2013, 24, 1840–1849. [Google Scholar] [CrossRef] [PubMed]
- Aslan, B.; Ozpolat, B.; Sood, A.K.; Lopez-Berestein, G. Nanotechnology in cancer therapy. J. Drug Target. 2013, 21, 904–913. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Jang, J. Antimicrobial polymer nanostructures: Synthetic route, mechanism of action and perspective. Adv. Colloid Interface Sci. 2014, 203, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, H.; Katsumata, R. Antibiotic resistance in bacteria and its future for novel antibiotic development. Biosci. Biotechnol. Biochem. 2006, 70, 1060–1075. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.D. Bacterial resistance and topical antimicrobial wash products. Am. J. Infect. Control 1999, 27, 351–363. [Google Scholar] [CrossRef] [PubMed]
- Poole, K. Mechanisms of bacterial biocide and antibiotic resistance. J. Appl. Microbiol. 2002, 92, 55S–64S. [Google Scholar] [CrossRef] [PubMed]
- Russell, A.D. Biocide use and antibiotic resistance: The relevance of laboratory findings to clinical and environmental situations. Lancet Infect. Dis. 2003, 3, 794–803. [Google Scholar] [CrossRef] [PubMed]
- Carmona-Ribeiro, A.M.; Carrasco, L.D.M. Cationic antimicrobial polymers and their assemblies. Int. J. Mol. Sci. 2013, 14, 9906–9946. [Google Scholar] [CrossRef] [PubMed]
- Sinha, R.; Roychoudhury, J.; Palit, P.; Ali, N. Cationic liposomal sodium stibogluconate (SSG), a potent therapeutic tool for treatment of infection by SSG-sensitive and -resistant Leishmania donovani. Antimicrob. Agents Chemother. 2015, 59, 344–355. [Google Scholar] [CrossRef] [PubMed]
- Carmona-Ribeiro, A.M.; Carrasco, L.D.M. Novel formulations for antimicrobial peptides. Int. J. Mol. Sci. 2014, 15, 18040–18083. [Google Scholar] [CrossRef] [PubMed]
- Domagk, G. Eineneueklasse von disinfektionsmitteln. Dtsch. Med. Wochenschr. 1935, 61, 829–832. [Google Scholar] [CrossRef]
- Ravikumar, T.; Murata, H.; Koepsel, R.R.; Russell, A.J. Surface-active antifungal polyquaternary amine. Biomacromolecules 2006, 7, 2762–2769. [Google Scholar] [CrossRef] [PubMed]
- Pasquier, N.; Keul, H.; Heine, E.; Moeller, M. From multifunctionalized poly(ethylene imine)s toward antimicrobial coatings. Biomacromolecules 2007, 8, 2874–2882. [Google Scholar] [CrossRef] [PubMed]
- Melo, L.D.; Mamizuka, E.M.; Carmona-Ribeiro, A.M. Antimicrobial particles from cationic lipid and polyelectrolytes. Langmuir 2010, 26, 12300–12306. [Google Scholar] [CrossRef] [PubMed]
- Melo, L.D.; Carmona-Ribeiro, A.M. Fungicidal nanoparticles of low toxicity from cationic lipid and polyelectrolytes. NSTI Nanotechnol. 2012, 3, 350–353. [Google Scholar]
- Song, F.; Li, X.; Wang, Q.; Liao, L.; Zhang, C. Nanocomposite hydrogels and their applications in drug delivery and tissue engineering. J. Biomed. Nanotechnol. 2015, 11, 40–52. [Google Scholar] [CrossRef]
- Zhan, Y.; Zeng, W.; Jiang, G.; Wang, Q.; Shi, X.; Zhou, Z.; Deng, H.; Du, Y. Construction of lysozyme exfoliated rectorite-based electrospun nanofibrous membranes for bacterial inhibition. J. Appl. Polym. Sci. 2015, 132, 41496–41505. [Google Scholar] [CrossRef]
- Ding, F.; Deng, H.; Du, Y.; Shi, X.; Wang, Q. Emerging chitin and chitosan nanofibrous materials for biomedical applications. Nanoscale 2014, 6, 9477–9493. [Google Scholar] [CrossRef] [PubMed]
- Carmona-Ribeiro, A.M.; Carrasco, L.D.M. Fungicidal assemblies and their mode of action. OA Biotechnol. 2013, 2, 25:1–25:8. [Google Scholar] [CrossRef]
- Vieira, D.B.; Carmona-Ribeiro, A.M. Cationic nanoparticles for delivery of amphotericin B: Preparation, characterization and activity in vitro. J. Nanobiotechnol. 2008, 6, 6:1–6:13. [Google Scholar] [CrossRef] [Green Version]
- Carmona-Ribeiro, A.M. Lipid bilayer fragments and disks in drug delivery. Curr. Med. Chem. 2006, 13, 1359–1370. [Google Scholar] [CrossRef] [PubMed]
- Carmona-Ribeiro, A.M. Biomimetic nanoparticles: Preparation, characterization and biomedical applications. Int. J. Nanomed. 2010, 5, 249–259. [Google Scholar] [CrossRef]
- Decher, G.; Hong, J.D.; Schmitt, J. Buildup of ultrathin multilayer films by a self-assembly process: III. Consecutively alternating adsorption of anionic and cationic polyelectrolytes on charged surfaces. Thin Solid Films 1992, 210–211, 831–835. [Google Scholar] [CrossRef]
- Carmona-Ribeiro, A.M. Preparation and characterization of biomimetic nanoparticles for drug delivery. Methods Mol. Biol. 2012, 906, 283–294. [Google Scholar] [PubMed]
- Martins, L.M.S.; Mamizuka, E.M.; Carmona-Ribeiro, A.M. Cationic vesicles as bactericides. Langmuir 1997, 13, 5583–5587. [Google Scholar] [CrossRef]
- Campanhã, M.T.N.; Mamizuka, E.M.; Carmona-Ribeiro, A.M. Interactions between cationic liposomes and bacteria: The physical-chemistry of the bactericidal action. J. Lipid Res. 1999, 40, 1495–1500. [Google Scholar] [PubMed]
- Campanhã, M.T.N.; Mamizuka, E.M.; Carmona-Ribeiro, A.M. Interactions between cationic vesicles and Candida albicans. J. Phys. Chem. B 2001, 105, 8230–8236. [Google Scholar] [CrossRef]
- Melo, L.D.; Palombo, R.R.; Petri, D.F.S.; Bruns, M.; Pereira, E.M.A.; Carmona-Ribeiro, A.M. Structure-activity relationship for quaternary ammonium compounds hybridized with poly (methyl methacrylate). ACS Appl. Mater. Interfaces 2011, 3, 1933–1939. [Google Scholar] [CrossRef] [PubMed]
- Barbassa, L.; Mamizuka, E.M.; Carmona-Ribeiro, A.M. Supramolecular assemblies of rifampicin and cationic bilayers: Preparation, characterization and micobactericidal activity. BMC Biotechnol. 2011, 11, 40:1–40:8. [Google Scholar] [CrossRef]
- Naves, A.F.; Palombo, R.R.; Carrasco, L.D.M.; Carmona-Ribeiro, A.M. Antimicrobial particles from emulsion polymerization of methyl methacrylate in the presence of quaternary ammonium surfactants. Langmuir 2013, 29, 9677–9684. [Google Scholar] [CrossRef] [PubMed]
- Lilly, H.A.; Lowbury, E.J.; Wilkins, M.D. Limits to progressive reduction of resident skin bacteria by disinfection. J. Clin. Pathol. 1979, 32, 382–385. [Google Scholar] [CrossRef] [PubMed]
- Veldhuizen, E.J.A.; Brouwer, E.C.; Schneider, V.A.F.; Fluit, A.C. Chicken cathelicidins display antimicrobial activity against multiresistant bacteria without inducing strong resistance. PLoS One 2013, 8, e61964. [Google Scholar] [CrossRef] [PubMed]
- Otto, M. Staphylococcus epidermidis—The “accidental” pathogen. Nat. Rev. Microbiol. 2009, 7, 555–567. [Google Scholar] [CrossRef] [PubMed]
- Merchat, M.; Bertolini, G.; Giacomini, P.; Villanueva, A.; Jori, G. Meso-substituted cationic porphyrins as efficient photosensitizers of gram-positive and gram-negative bacteria. J. Photochem. Photobiol. B 1996, 32, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Nitzan, Y.; Gutterman, M.; Malik, Z.; Ehrenberg, B. Inactivation of gram-negative bacteria by photosensitized porphyrins. Photochem. Photobiol. 1992, 55, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Salton, M.R.J. Lytic agents, cell permeability and monolayer penetrability. J. Gen. Physiol. 1968, 52, 227–252. [Google Scholar] [CrossRef] [PubMed]
- Denyer, S.P. Mechanisms of action of antibacterial biocides. Int. Biodeterior. Biodegrad. 1995, 36, 227–245. [Google Scholar] [CrossRef]
- Jeon, S.J.; Oh, M.; Yeo, W.S.; Galvão, K.N.; Jeong, K.C. Underlying mechanism of antimicrobial activity of chitosan microparticles and implications for the treatment of infectious diseases. PLoS One 2014, 9, e92723. [Google Scholar] [CrossRef] [PubMed]
- Calabretta, M.K.; Kumar, A.; McDermott, A.M.; Cai, C. Antibacterial activities of poly (amidoamine) dendrimers terminated with amino and poly (ethylene glycol) groups. Biomacromolecules 2007, 8, 1807–1811. [Google Scholar] [CrossRef]
- Devarakonda, B.; Judefeind, A.; Chigurupati, S.; Thomas, S.; Shah, G.V.; Otto, D.P.; de Villiers, M.M. The effect of polyamidoamine dendrimers on the in vitro cytotoxicity of paclitaxel in cultured prostate cancer (PC-3M) cells. J. Biomed. Nanotechnol. 2007, 3, 384–393. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, Z.J.; Zhu, J.S.; Tang, Y.L.; Canady, T.D.; Chi, E.Y.; Schanze, K.S.; Whitten, D.G. Dark antimicrobial mechanisms of cationic phenylene ethynylene polymers and oligomers against Escherichia coli. Polymers 2011, 3, 1199–1214. [Google Scholar] [CrossRef]
- Veiga, A.S.; Sinthuvanich, C.; Gaspar, D.; Franquelim, H.G.; Castanho, M.A.; Schneider, J.P. Arginine-rich self-assembling peptides as potent antibacterial gels. Biomaterials 2012, 33, 8907–8916. [Google Scholar] [CrossRef] [PubMed]
- Strömstedt, A.A.; Ringstad, L.; Schmidtchen, A.; Malmsten, M. Interaction between amphiphilic peptides and phospholipid membranes. Curr. Opin. Colloid Interface Sci. 2010, 15, 467–478. [Google Scholar] [CrossRef]
- Pasupuleti, M.; Schmidtchen, A.; Malmsten, M. Antimicrobial peptides: Key components of the innate immune system. Crit. Rev. Biotechnol. 2012, 32, 143–171. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhang, Y. Bacteria-removing and bactericidal efficiencies of pdadmac composite coagulants in enhanced coagulation treatment. Clean 2013, 41, 37–42. [Google Scholar]
- Sicchierolli, S.M.; Mamizuka, E.M.; Carmona-Ribeiro, A.M. Bacteria flocculation and death by cationic vesicles. Langmuir 1995, 11, 2991–2995. [Google Scholar] [CrossRef]
- Tiller, J.C.; Liao, C.J.; Lewis, K.; Klibanov, A.M. Designing surfaces that kill bacteria on contact. Proc. Natl. Acad. Sci. USA 2001, 98, 5981–5985. [Google Scholar] [CrossRef] [PubMed]
- Milovic, N.M.; Wang, J.; Lewis, K.; Klibanov, A.M. Immobilized N-alkylated polyethylenimine avidly kills bacteria by rupturing cell membranes with no resistance developed. Biotechnol. Bioeng. 2005, 90, 715–722. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Poon, Y.F.; Li, W.; Zhu, H.Y.; Yeap, S.H.; Cao, Y.; Qi, X.; Zhou, C.; Lamrani, M.; Beuerman, R.W.; et al. A polycationic antimicrobial and biocompatible hydrogel with microbe membrane suctioning ability. Nat. Mater. 2011, 10, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Ling, L.L.; Schneider, T.; Peoples, A.J.; Spoering, A.L.; Engels, I.; Conlon, B.P.; Mueller, A.; Schäberle, T.F.; Hughes, D.E.; Epstein, S.; et al. A new antibiotic kills pathogens without detectable resistance. Nature 2015, 517, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Lei, B.; Li, P.; Ma, P.X. Functionalized scaffolds to enhance tissue regeneration. Regen. Biomater. 2015, 2, 1–11. [Google Scholar] [CrossRef]
- Nizet, V. Antimicrobial peptide resistance mechanisms of human bacterial pathogens. Curr. Issues Mol. Biol. 2006, 8, 11–26. [Google Scholar] [PubMed]
- Gregoriadis, G. Engineering liposomes for drug delivery: Progress and problems. Trends Biotechnol. 1995, 13, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Lincopan, N.; Mamizuka, E.M.; Carmona-Ribeiro, A.M. In vivo activity of a novel amphotericin B formulation with synthetic cationic bilayer fragments. J. Antimicrob. Chemother. 2003, 52, 412–418. [Google Scholar] [CrossRef] [PubMed]
- Lincopan, N.; Espíndola, N.M.; Vaz, A.J.; da Costa, M.H.; Faquim-Mauro, E.; Carmona-Ribeiro, A.M. Novel immunoadjuvants based on cationic lipid: Preparation, characterization and activity in vivo. Vaccine 2009, 27, 5760–5771. [Google Scholar] [CrossRef] [PubMed]
- Rozenfeld, J.H.; Silva, S.R.; Ranéia, P.A.; Faquim-Mauro, E.; Carmona-Ribeiro, A.M. Stable assemblies of cationic bilayer fragments and CpG oligonucleotide with enhanced immunoadjuvant activity in vivo. J. Control. Release 2012, 160, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Carmona-Ribeiro, A.M. Cationic nanostructures for vaccines. In Immune Response Activation, 1st ed.; Duc, G.H.T., Ed.; InTech: Rijeka, Croatia, 2014; pp. 3–43. [Google Scholar]
- Carvalho, L.A.; Carmona-Ribeiro, A.M. Interactions between cationic vesicles and serum proteins. Langmuir 1998, 14, 6077–6081. [Google Scholar] [CrossRef]
- Carmona-Ribeiro, A.M. Synthetic amphiphile vesicles. Chem. Soc. Rev. 1992, 21, 209–214. [Google Scholar] [CrossRef]
- Grabowski, E.; Morrison, I. Particle size distribution from analysis of quasielastic light scattering data. In Measurements of Suspended Particles by Quasielastic Light Scattering; Dahneke, B., Ed.; Wiley-Interscience: New York, NY, USA, 1983; pp. 199–236. [Google Scholar]
- Chapin, K.C.; Lauderdale, T. Reagents, stains, and media: Bacteriology. In Manual of Clinical Microbiology, 9th ed.; Murray, P.R., Baron, E.J., Jorgensen, J.H., Landry, M.L., Pfaller, M.A., Eds.; ASM Press: Washington, DC, USA, 2007; pp. 334–364. [Google Scholar]
- Hoque, J.; Akkapeddi, P.; Yarlagadda, V.; Uppu, D.S.S.M.; Kumar, P.; Haldar, J. Cleavable cationic antibacterial amphiphiles: Synthesis, mechanism of action, and cytotoxicities. Langmuir 2012, 28, 12225–12234. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Melo Carrasco, L.D.; Sampaio, J.L.M.; Carmona-Ribeiro, A.M. Supramolecular Cationic Assemblies against Multidrug-Resistant Microorganisms: Activity and Mechanism of Action. Int. J. Mol. Sci. 2015, 16, 6337-6352. https://doi.org/10.3390/ijms16036337
De Melo Carrasco LD, Sampaio JLM, Carmona-Ribeiro AM. Supramolecular Cationic Assemblies against Multidrug-Resistant Microorganisms: Activity and Mechanism of Action. International Journal of Molecular Sciences. 2015; 16(3):6337-6352. https://doi.org/10.3390/ijms16036337
Chicago/Turabian StyleDe Melo Carrasco, Letícia Dias, Jorge Luiz Mello Sampaio, and Ana Maria Carmona-Ribeiro. 2015. "Supramolecular Cationic Assemblies against Multidrug-Resistant Microorganisms: Activity and Mechanism of Action" International Journal of Molecular Sciences 16, no. 3: 6337-6352. https://doi.org/10.3390/ijms16036337