Novel Cholesterol-Based Cationic Lipids as Transfecting Agents of DNA for Efficient Gene Delivery
Abstract
:1. Introduction

2. Results and Discussion
2.1. Chemistry

2.2. Characterization of Neat Cationic Liposomes
| Cationic Lipid | Size | Polydispersity Index (PDI) | Zeta Potential |
|---|---|---|---|
| 1a | 143.0 ± 1.7 nm | 0.260 ± 0.020 | 23.7 ± 3.9 mV |
| 1b | 144.5 ± 2.0 nm | 0.289 ± 0.008 | 47.7 ± 4.1 mV |

2.3. Gel Electrophoresis

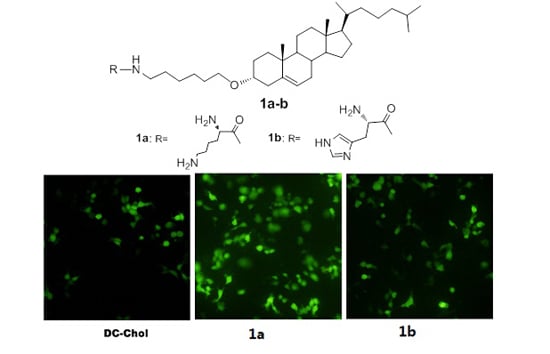
2.4. Transfection Biology
2.4.1. Optimization of the Ratio of Lipid and DOPE


2.4.2. Optimization of N/P Charge Ratio
2.5. Cytotoxicity Assay



3. Experimental Section
3.1. Materials
3.2. Synthesis of Cationic Lipids
3.2.1. Cholest-5-en-3β-tosylate (2)
3.2.2. Cholest-5-en-3β-hexane (3)
3.2.3. Cholest-5-en-3β-yl 6-Aminohexyl Ether (4)
3.2.4. General Method for Synthesis of the Lipids 1a and 1b
3.3. Preparation of Liposomes
3.4. Size Measurement and Zeta Potential Analysis of Liposome
3.5. Gel Retardation Assay
3.6. Cell Culture
3.7. In Vitro Transfection
3.7.1. Fluorescence Microscopy
3.7.2. Flow Cytometry
3.8. Cytotoxicity
3.9. Statistical Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Junquera, E.; Aicart, E. Cationic lipids as transfecting agents of DNA in gene therapy. Curr. Top. Med. Chem. 2014, 14, 649–663. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Zeng, X.; Liu, M.; Deng, Y.; He, N. Current progress in gene delivery technology based on chemical methods and nano-carriers. Theranostics 2014, 4, 240–255. [Google Scholar] [CrossRef] [PubMed]
- Anderson, W.F. Human gene therapy. Nature 1998, 392, 25–30. [Google Scholar]
- Islam, M.A.; Firdous, J.; Choi, Y.J.; Yun, C.H.; Cho, C.S. Regulation of endocytosis by non-viral vectors for efficient gene activity. J. Biomed. Nanotechnol. 2014, 10, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Ronsin, G.; Perrin, C.; Guedat, P.; Kremer, A.; Camilleri, P.; Kirby, A.J. Novel spermine-based cationic gemini surfactants for gene delivery. Chem. Commun. 2001, 21, 2234–2235. [Google Scholar] [CrossRef]
- Raper, S.E.; Yudkoff, M.; Chirmule, N.; Gao, G.P.; Nunes, F.; Haskal, Z.J.; Furth, E.E.; Propert, K.J.; Robinson, M.B.; Magosin, S.; et al. A pilot study of in vivo liver-directed gene transfer with an adenoviral vector in partial ornithine transcarbamylase deficiency. Hum. Gene. Ther. 2002, 13, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Wiethoff, C.M.; Koe, J.G.; Koe, G.S.; Middaugh, C.R. Compositional effects of cationic lipid/DNA delivery systems on transgene expression in cell culture. J. Pharm. Sci. 2004, 93, 108–123. [Google Scholar] [CrossRef] [PubMed]
- Marshall, E. Gene therapy. What to do when clear success comes with an unclear risk? Science 2002, 298, 510–511. [Google Scholar] [CrossRef] [PubMed]
- Mintzer, M.A.; Simanek, E.E. Nonviral vectors for gene delivery. Chem. Rev. 2009, 109, 259–302. [Google Scholar] [CrossRef] [PubMed]
- Sen, J.; Chaudhuri, A. Design, syntheses, and transfection biology of novel non-cholesterol-based guanidinylated cationic lipids. J. Med. Chem. 2005, 48, 812–820. [Google Scholar] [CrossRef] [PubMed]
- Suh, M.S.; Shim, G.; Lee, H.Y.; Han, S.E.; Yu, Y.H.; Choi, Y.; Kim, K.; Kwon, I.C.; Weon, K.Y.; Kim, Y.B.; Oh, Y.K. Anionic amino acid-derived cationic lipid for siRNA delivery. J. Control. Release 2009, 140, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Song, H.; Luo, K.; He, B.; Nie, Y.; Yang, Y.; Wu, Y.; Gu, Z. Gene transfer efficacies of serum-resistant amino acids-based cationic lipids: Dependence on headgroup, lipoplex stability and cellular uptake. Int. J. Pharm. 2011, 408, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Sheng, R.; Luo, T.; Li, H.; Sun, J.; Wang, Z.; Cao, A. “Click” synthesized sterol-based cationic lipids as gene carriers, and the effect of skeletons and headgroups on gene delivery. Bioorg. Med. Chem. 2013, 21, 6366–6377. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, A.; Kondiah, P.; Bhattacharya, S. Design, synthesis, and in vitro gene delivery efficacies of novel cholesterol-based gemini cationic lipids and their serum compatibility: A structure-activity investigation. J. Med. Chem. 2007, 50, 2432–2442. [Google Scholar] [CrossRef] [PubMed]
- Chien, P.Y.; Wang, J.; Carbonaro, D.; Lei, S.; Miller, B.; Sheikh, S.; Ali, S.M.; Ahmad, M.U.; Ahmad, I. Novel cationic cardiolipin analogue-based liposome for efficient DNA and small interfering RNA delivery in vitro and in vivo. Cancer Gene Ther. 2005, 12, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Lv, H.; Zhang, S.; Wang, B.; Cui, S.; Yan, J. Toxicity of cationic lipids and cationic polymers in gene delivery. J. Control. Release 2006, 114, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Pungente, M.D.; Jubeli, E.; Opstad, C.L.; Al-Kawaz, M.; Barakat, N.; Ibrahim, T.; Abdul, K.N.; Raju, L.; Jones, R.; Leopold, P.L.; et al. Synthesis and preliminary investigations of the siRNA delivery potential of novel, single-chain rigid cationic carotenoid lipids. Molecules 2012, 17, 3484–3500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Egilmez, N.K.; Iwanuma, Y.; Bankert, R.B. Evaluation and optimization of different cationic liposome formulations for in vivo gene transfer. Biochem. Biophys. Res. Commun. 1996, 221, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Farhood, H.; Gao, X.; Barsoum, J.; Huang, L. Codelivery to mammalian cells of a transcriptional factor with cis-acting element using cationic liposomes. Anal. Biochem. 1995, 225, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Huang, L. A novel cationic liposome reagent for efficient transfection of mammalian cells. Biochem. Biophys. Res. Commun. 1991, 179, 280–285. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.K.; Seu, Y.B.; Bae, Y.U.; Kwak, T.W.; Kang, H.; Moon, I.J.; Hwang, G.B.; Park, S.Y.; Doh, K.O. Efficient delivery of plasmid DNA using cholesterol-based cationic lipids containing polyamines and ether linkages. Int. J. Mol. Sci. 2014, 15, 7293–7312. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.K.; Doh, K.O.; Nam, J.H.; Kang, H.; Park, J.G.; Moon, I.J.; Seu, Y.B. Synthesis of novel cholesterol-based cationic lipids for gene delivery. Bioorg. Med. Chem. Lett. 2009, 19, 2986–2989. [Google Scholar] [CrossRef] [PubMed]
- Okuda, T.; Sugiyama, A.; Niidome, T.; Aoyagi, H. Characters of dendritic poly(l-lysine) analogues with the terminal lysines replaced with arginines and histidines as gene carriers in vitro. Biomaterials 2004, 25, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Karmali, P.P.; Kumar, V.V.; Chaudhuri, A. Design, syntheses and in vitro gene delivery efficacies of novel mono-, di- and trilysinated cationic lipids: A structure-activity investigation. J. Med. Chem. 2004, 47, 2123–2132. [Google Scholar] [CrossRef] [PubMed]
- Karmali, P.P.; Majeti, B.K.; Sreedhar, B.; Chaudhuri, A. In vitro gene transfer efficacies and serum compatibility profiles of novel mono-, di-, and tri-histidinylated cationic transfection lipids: A structure-activity investigation. Bioconjug. Chem. 2006, 17, 159–171. [Google Scholar] [CrossRef] [PubMed]
- Shigeta, K.; Kawakami, S.; Higuchi, Y.; Okuda, T.; Yagi, H.; Yamashita, F.; Hashida, M. Novel histidine-conjugated galactosylated cationic liposomes for efficient hepatocyte-selective gene transfer in human hepatoma HepG2 cells. J. Control. Release 2007, 118, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.K.; Mani, P.; Sharma, N.R.; Krishnan, A.; Kumar, V.V.; Reddy, B.S.; Chaudhuri, A.; Roy, R.P.; Sarkar, D.P. Histidylated lipid-modified Sendai viral envelopes mediate enhanced membrane fusion and potentiate targeted gene delivery. J. Biol. Chem. 2005, 280, 35399–35409. [Google Scholar] [CrossRef] [PubMed]
- Sheng, R.; Luo, T.; Li, H.; Sun, J.; Wang, Z.; Cao, A. Cholesterol-based cationic lipids for gene delivery: contribution of molecular structure factors to physico-chemical and biological properties. Colloids Surf. B Biointerfaces 2014, 116, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, Y.K.; Visweswariah, S.S.; Bhattacharya, S. Advantage of the ether linkage between the positive charge and the cholesteryl skeleton in cholesterol-based amphiphiles as vectors for gene delivery. Bioconjug. Chem. 2002, 13, 378–384. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, Y.K.; Visweswariah, S.S.; Bhattacharya, S. Nature of linkage between the cationic headgroup and cholesteryl skeleton controls gene transfection efficiency. Febs. Lett. 2000, 473, 341–344. [Google Scholar] [CrossRef] [PubMed]
- Angelini, G.; Pisani, M.; Mobbili, G.; Marini, M.; Gasbarri, C. Neutral liposomes containing crown ether-lipids as potential DNA vectors. Biochim. Biophys. Acta 2013, 1828, 2506–2512. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.S.; Lee, E.J.; Jang, H.S.; Park, J.S. New cationic liposomes for gene transfer into mammalian cells with high efficiency and low toxicity. Bioconjug. Chem. 2001, 12, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Hu, J.; Qiao, W.; Li, Z.; Zhan, S.; Cheng, L. Synthesis and characterization of a series of carbamate-linked cationic lipids for gene delivery. Lipids 2005, 40, 839–848. [Google Scholar] [CrossRef] [PubMed]
- Felgner, J.H.; Kumar, R.; Sridhar, C.N.; Wheeler, C.J.; Tsai, Y.J.; Border, R.; Ramsey, P.; Martin, M.; Felgner, P.L. Enhanced gene delivery and mechanism studies with a novel series of cationic lipid formulations. J. Biol. Chem. 1994, 269, 2550–2561. [Google Scholar] [PubMed]
- Zhang, Y.; Li, H.; Sun, J.; Gao, J.; Liu, W.; Li, B.; Guo, Y.; Chen, J. DC-Chol/DOPE cationic liposomes: A comparative study of the influence factors on plasmid pDNA and siRNA gene delivery. Int. J. Pharm. 2010, 390, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, A.; Aziz, S.A.; Gilbert, M.; Werner, D.; Noe, C.R. Synthesis of cholesterol modified cationic lipids for liposomal drug delivery of antisense oligonucleotides. Eur. J. Pharm. Biopharm. 1999, 47, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.B.; Nielsen, S.E.; Berg, K. Re-examination and further development of a precise and rapid dye method for measuring cell growth/cell kill. J. Immunol. Methods 1989, 119, 203–210. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ju, J.; Huan, M.-L.; Wan, N.; Qiu, H.; Zhou, S.-Y.; Zhang, B.-L. Novel Cholesterol-Based Cationic Lipids as Transfecting Agents of DNA for Efficient Gene Delivery. Int. J. Mol. Sci. 2015, 16, 5666-5681. https://doi.org/10.3390/ijms16035666
Ju J, Huan M-L, Wan N, Qiu H, Zhou S-Y, Zhang B-L. Novel Cholesterol-Based Cationic Lipids as Transfecting Agents of DNA for Efficient Gene Delivery. International Journal of Molecular Sciences. 2015; 16(3):5666-5681. https://doi.org/10.3390/ijms16035666
Chicago/Turabian StyleJu, Jia, Meng-Lei Huan, Ning Wan, Hai Qiu, Si-Yuan Zhou, and Bang-Le Zhang. 2015. "Novel Cholesterol-Based Cationic Lipids as Transfecting Agents of DNA for Efficient Gene Delivery" International Journal of Molecular Sciences 16, no. 3: 5666-5681. https://doi.org/10.3390/ijms16035666