Neuritogenic and Neuroprotective Properties of Peptide Agonists of the Fibroblast Growth Factor Receptor
Abstract
:1. Introduction
1.1. FGF Receptors
1.2. Ligands of FGF Receptors
1.2.1. FGFs
1.2.2. Cell Adhesion Molecules
2. Peptide Agonists of FGFR Derived from FGFs
2.1. Canofins: Peptide Agonists of FGFR Derived from the Canonical FGFR Binding Sites
2.2. Hexafins: Peptide Agonists of FGFR Derived from the β6–β7 Loop Regions of Various FGFs
2.3. Dekafins: Peptide Agonists of FGFR Derived from the β10–β11 Loop Regions of Various FGFs
3. Peptide Agonists of FGFR Derived from NCAM
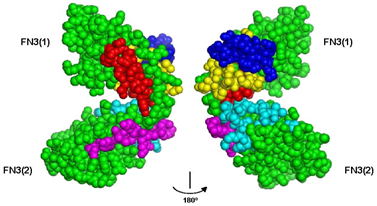
3.1. Peptide Agonists of FGFR Derived from the First NCAM FN3 Module
3.2. Peptide Agonists of FGFR Derived from the Second NCAM FN3 Module
4. Concluding Remarks
Acknowledgments
References
- Powers, CJ; McLeskey, SW; Wellstein, A. Fibroblast growth factors, and their receptors and signaling. Endocr. Relat. Cancer 2000, 7, 165–197. [Google Scholar]
- Goldstrohm, AC; Greenleaf, AL; Garcia-Blanco, MA. Co-transcriptional splicing of pre-messenger RNAs: Considerations for the mechanism of alternative splicing. Gene 2001, 277, 31–47. [Google Scholar]
- Orniz, DM. FGFs, heparan sulfate and FGFRs: Complex interactions essential for development. Bioessays 2000, 22, 108–112. [Google Scholar]
- Yeh, BK; Igarashi, M; Eliseenkova, AV; Plotnikov, AN; Sher, I; Ron, D; Aaronson, SA; Mohammadi, M. Structural basis by which alternative splicing confers specificity in fibroblast growth factor receptors. Proc. Natl. Acad. Sci. USA 2003, 100, 2266–2271. [Google Scholar]
- Mohammadi, M; Olsen, SK; Ibrahimi, OA. Structural basis for fibroblast growth factor receptor activation. Cytokine Growth Factor Rev 2005, 16, 107–137. [Google Scholar]
- Eswarakumar, VP; Lax, I; Schlessinger, J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev 2005, 16, 139–149. [Google Scholar]
- Dailey, L; Ambrosetti, D; Mansukhani, A; Basilico, C. Mechanisms underlying differential responses to FGF signaling. Cytokine Growth Factor Rev 2005, 16, 233–247. [Google Scholar]
- Zhang, J; Cousens, LS; Barr, PJ; Sprang, SR. Three-dimensional structure of human basic fibroblast growth factor, a structural homolog of interleukin 1β. Proc. Natl. Acad. Sci. USA 1991, 88, 3446–3450. [Google Scholar]
- Burgess, WH; Maciag, T. The heparin-binding (fibroblast) growth factor family of protein. Ann. Rev. Biochem 1989, 58, 575–606. [Google Scholar]
- Zhu, X; Komiya, H; Chirino, A; Faham, S; Fox, GM; Arakawa, T; Hsu, BT; Rees, DC. Three-dimensional structures of acidic and basic fibroblast growth factors. Science 1991, 251, 90–93. [Google Scholar]
- Ornitz, DM; Itoh, N. Fibroblast growth factors. Genome Biol 2001, 2, 3005:1–3005:12. [Google Scholar]
- Ornitz, DM; Xu, J; Colvin, JS; McEwen, DG; MacArthur, CA; Coulier, F; Gao, G; Goldfarb, M. Receptor specificity of the fibroblast growth factor family. J. Biol. Chem 1996, 271, 15292–15297. [Google Scholar]
- Miller, DL; Ortega, S; Bashayan, O; Basch, R; Basilico, C. Compensation by fibroblast growth factor 1 (FGF1) does not account for the mild phenotypic defects observed in FGF2 null mice. Mol. Cell Biol 2000, 20, 2260–2268. [Google Scholar]
- Raballo, R; Rhee, J; Lyn-Cook, R; Leckman, JF; Schwartz, ML; Vaccarino, FM. Basic fibroblast growth factor (Fgf2) is necessary for cell proliferation and neurogenesis in the developing cerebral cortex. J. Neurosci 2000, 20, 5012–5023. [Google Scholar]
- Reuss, B; von Bohlen und Halbach, O. Fibroblast growth factors and their receptors in the central nervous system. Cell Tissue Res 2003, 313, 139–157. [Google Scholar]
- Fiore, F; Sebille, A; Birnbaum, D. Skeletal muscle regeneration is not impaired in Fgf6 -/- mutant mice. Biochem. Biophys. Res. Commun 2000, 272, 138–143. [Google Scholar]
- Reifers, F; Bohli, H; Walsh, EC; Crossley, PH; Stainier, DY; Brand, M. Fgf8 is mutated in zebrafish acerebellar (ace) mutants and is required for maintenance of midbrain-hindbrain boundary development and somitogenesis. Development 1998, 125, 2381–2395. [Google Scholar]
- Xu, J; Liu, Z; Ortitz, DM. Temporal and spatial gradients of Fgf8 and Fgf17 regulate proliferation and differentiation of midline cerebellar structures. Development 2000, 127, 1833–1843. [Google Scholar]
- Grieshammer, U; Cebrian, C; Ilagan, R; Meyers, E; Herzlinger, D; Martin, GR. FGF8 is required for cell survival at distinct stages of nephrogenesis and for regulation of gene expression in nascent nephrons. Development 2005, 132, 3847–3857. [Google Scholar]
- Garces, A; Nishimune, H; Philippe, JM; Pettmann, B; deLapeyriere, O. FGF9: A motoneuron survival factor expressed by medial thoratic and sacral motoneurons. J. Neurosci. Res 2000, 60, 1–9. [Google Scholar]
- Kinkl, N; Ruiz, J; Vecino, E; Frasson, M; Sahel, J; Hicks, D. Possible involvement of a fibroblast growth factor 9 (FGF9)-FGF receptor-3-mediated pathway in adult pig retinal ganglion cell survival in vitro. Mol. Cell Neurosci 2003, 23, 39–53. [Google Scholar]
- Saffell, L; Williams, EJ; Mason, IJ; Walsh, FS; Doherty, P. Expression of a dominant negative FGF receptor inhibits axonal growth and FGF receptor phosphorylation stimulated by CAMs. Neuron 1997, 18, 231–242. [Google Scholar]
- Kulahin, N; Li, S; Hinsby, A; Kiselyov, V; Berezin, V; Bock, E. Fibronectin type III (FN3) modules of the neuronal cell adhesion molecule L1 interact directly with the fibroblast growth factor (FGF) receptor. Mol. Cell Neurosci 2008, 37, 528–536. [Google Scholar]
- Kirschbaum, K; Kriebel, M; Kranz, EU; Pötz, O; Volkmer, H. Analysis of non-canonical fibroblast growth factor receptor 1 (FGFR1) interaction reveals regulatory and activating domains of neurofascin. J. Biol. Chem 2009, 42, 28533–28542. [Google Scholar]
- Owczarek, S; Kiryushko, D; Larsen, MH; Kastrup, JS; Gajhede, M; Sandi, C; Berezin, V; Bock, E; Soroka, V. Neuroplastin-55 binds to and signals through the fibroblast growth factor receptor. FASEB J 2010, 24, 1139–1150. [Google Scholar]
- Doherty, P; Walsh, FS. Signal transduction events underlying neurite outgrowth stimulated by cell adhesion molecules. Curr. Opin. Neurobiol 1994, 4, 49–55. [Google Scholar]
- Williams, EJ; Walsh, FS; Doherty, P. Tyrosine kinase inhibitors can differentially inhibit integrin-dependent and CAM stimulated neurite outgrowth. J. Cell Biol 1994a, 124, 1029–1037. [Google Scholar]
- Williams, EJ; Furness, J; Walsh, FS; Doherty, P. Characterisation of the second messenger pathway underlying neurite outgrowth stimulated by FGF. Development 1994b, 120, 1685–1693. [Google Scholar]
- Williams, EJ; Furness, J; Walsh, FS; Doherty, P. Activation of the FGF receptor underlies neurite outgrowth stimulated by L1, N-CAM, and N-cadherin. Neuron 1994c, 13, 583–594. [Google Scholar]
- Saffell, L; Walsh, FS; Doherty, P. Expression of NCAM containing VASE in neurons can account for a developmental loss in their neurite outgrowth response to NCAM in a cellular substratum. J. Cell Biol 1994, 125, 427–436. [Google Scholar]
- Rønn, LC; Hartz, BP; Bock, E. The neural cell adhesion molecule (NCAM) in development and plasticity of the nervous system. Exp. Gerontol 1998, 33, 853–864. [Google Scholar]
- Rønn, LC; Doherty, P; Holm, A; Berezin, V; Bock, E. Neurite outgrowth induced by a synthetic peptide ligand of neural cell adhesion molecule requires fibroblast growth factor receptor activation. J. Neurochem 2000, 75, 665–671. [Google Scholar]
- Berezin, V; Bock, E; Poulsen, F. The neural cell adhesion molecule NCAM. Curr. Opin. Drug. Disc. Dev 2000, 3, 605–609. [Google Scholar]
- Hinsby, AM; Berezin, V; Bock, E. Molecular mechanisms of NCAM function. Front. Biosci 2004, 9, 2227–2244. [Google Scholar]
- Cremer, H; Chazal, G; Carleton, A; Goridis, C; Vincent, JD; Lledo, PM. Long-term but not short-term plasticity at mossy fiber synapses is impaired in neural cell adhesion molecule-deficient mice. Proc. Natl Acad. Sci. USA 1998, 95, 13242–13247. [Google Scholar]
- Cambon, K; Hansen, SM; Venero, C; Herrero, AI; Skibo, G; Berezin, V; Bock, E; Sandi, C. A synthetic neural cell adhesion molecule mimetic peptide promotes synaptogenesis, enhances presynaptic function, and facilitates memory consolidation. J. Neurosci 2004, 24, 4197–4204. [Google Scholar]
- Sandi, C. Stress, cognitive impairment and cell adhesion molecules. Nat. Rev. Neurosci 2004, 5, 917–930. [Google Scholar]
- Cremer, H; Chazal, G; Goridis, C; Represa, A. NCAM is essential for axonal growth and fasciculation in the hippocampus. Mol. Cell Neurosci 1997, 8, 323–335. [Google Scholar]
- Muller, D; Wang, C; Skibo, G; Toni, N; Cremer, H; Calaora, V; Rougon, G; Kiss, JZ. PSA-NCAM is required for activity-induced synaptic plasticity. Neuron 1996, 17, 413–422. [Google Scholar]
- Rutishauser, U. Polysialic acid in the plasticity of the developing and adult vertebrate nervous system. Nat. Rev. Neurosci 2008, 9, 26–35. [Google Scholar]
- Muller, D; Djebbara-Hannas, Z; Jourdain, P; Vutskits, L; Durbec, P; Rougon, G; Kiss, JZ. Brain-derived neurotrophic factor restores long-term potentiation in polysialic acid-neural cell adhesion molecule-deficient hippocampus. Proc. Natl. Acad. Sci. USA 2000, 97, 4315–4320. [Google Scholar]
- Jørgensen, OS; Bock, E. Brain specific synaptosomal membrane proteins demonstrated by crossed immunoelectrophoresis. J. Neurochem 1974, 23, 879–880. [Google Scholar]
- Walmod, PS; Kolkova, K; Berezin, V; Bock, E. Zippers make signals: NCAM-mediated molecular interactions and signal transduction. Neurochem. Res 2004, 29, 2015–2035. [Google Scholar]
- Thomsen, NK; Soroka, V; Jensen, PH; Berezin, V; Kiselyov, VV; Bock, E; Poulsen, FM. The three-dimensional structure of the first domain of neural cell adhesion molecule. Nat. Struct. Biol 1996, 3, 581–585. [Google Scholar]
- Kiselyov, VV; Skladchikova, G; Hinsby, AM; Jensen, PH; Kulahin, N; Soroka, V; Pedersen, N; Tsetlin, V; Poulsen, FM; Berezin, V; Bock, E. Structural basis for a direct interaction between FGFR1 and NCAM and evidence for a regulatory role of ATP. Structure 2003, 11, 691–701. [Google Scholar]
- Carafoli, F; Saffell, JL; Hohenester, E. Structure of the tandem fibronectin type 3 domains of neural cell adhesion molecule. J. Mol. Biol 2008, 2, 524–534. [Google Scholar]
- Frei, T; von Bohlen und Halbach, F; Wille, W; Schachner, M. Different extracellular domains of the neural cell adhesion molecule (N-CAM) are involved in different functions. J. Cell Biol 1992, 118, 177–194. [Google Scholar]
- Kasper, C; Rasmussen, H; Kastrup, JS; Ikemizu, S; Jones, EY; Berezin, V; Bock, E; Larsen, IK. Structural basis of cell-cell adhesion by NCAM. Nat. Struct. Biol 2000, 7, 389–393. [Google Scholar]
- Plotnikov, AN; Schlessinger, J; Hubbard, SR; Mohammadi, M. Structural basis for FGF receptor dimerization and activation. Cell 1999, 98, 641–650. [Google Scholar]
- Manfe, V; Kochoyan, A; Bock, E; Berezin, V. Peptides derived from specific interaction sites of the fibroblast growth factor (FGF) 2-FGF receptor complexes induce receptor activation and signalling. J Neurochem 2010, in press. [Google Scholar]
- Li, S; Christensen, C; Køhler, LB; Kiselyov, VV; Berezin, V; Bock, E. Agonists of fibroblast growth factor receptor induce neurite outgrowth and survival of cerebellar granule neurons. Dev. Neurobiol 2009, 69, 837–854. [Google Scholar]
- Rudenko, O; Tkach, V; Berezin, V; Bock, E. Effects of FGF receptor peptide agonists on animal behavior under normal and pathological conditions. Neurosci Res 2010, in press. [Google Scholar]
- Li, S; Christensen, C; Kiselyov, VV; Køhler, LB; Bock, E; Berezin, V. Fibroblast growth factor-derived peptides: Functional agonists of the fibroblast growth factor receptor. J. Neurochem 2008, 104, 667–682. [Google Scholar]
- Christensen, C; Lauridsen, JB; Berezin, V; Bock, E; Kiselyov, VV. The neural cell adhesion molecule binds to fibroblast growth factor receptor 2. FEBS Lett 2006, 580, 3386–3390. [Google Scholar]
- Hansen, SM; Køhler, LB; Li, S; Kiselyov, V; Christensen, C; Owczarek, S; Bock, E; Berezin, V. NCAM-derived peptides function as agonist for the firoblast growth factor receptor. J. Neurochem 2008, 106, 2030–2041. [Google Scholar]
- Anderson, AA; Kendal, CE; Garcia-Maya, M; Kenny, AV; Morris-Triggs, SA; Wu, T; Reynolds, R; Hohenester, E; Saffell, JL. A peptide from the first fibronectin domain of NCAM acts as an inverse agonist and stimulates FGF receptor activation, neurite outgrowth and survival. J. Neurochem 2005, 95, 570–583. [Google Scholar]
- Chen, Y; Li, S; Berezin, V; Bock, E. The fibroblast growth factor receptor (FGFR) agonist FGF1 and the neural cell adhesion molecule-derived peptide FGL activate FGFR substrate 2α differently. J. Neurosci. Res 2010, 88, 1882–1889. [Google Scholar]
- Neiiendam, JL; Kohler, LB; Christensen, C; Li, S; Pedersen, MV; Ditlevsen, DK; Kornum, MK; Kiselyov, VV; Berezin, V; Bock, E. An NCAM-derived FGF-receptor agonist, the FGL-peptide, induces neurite outgrowth and neuronal survival in primary rat neurons. J. Neurochem 2004, 91, 920–935. [Google Scholar]
- Downer, EJ; Cowley, TR; Cox, F; Maher, FO; Berezin, V; Bock, E; Lynch, MA. A synthetic NCAM-derived mimetic peptide, FGL, exerts anti-inflammatory properties via IGF-1 and interferon-γ modulation. J. Neurochem 2009, 109, 1516–1525. [Google Scholar]
- Secher, T; Novitskaia, V; Berezin, V; Bock, E; Glenthoj, B; Klementiev, B. A neural cell adhesion molecule-derived fibroblast growth factor receptor agonist, the FGL-peptide, promotes early postnatal sensorimotor development and enhances social memory retention. Neuroscience 2006, 141, 1289–1299. [Google Scholar]
- Skibo, GG; Lushnikova, IV; Voronin, KY; Dmitrieva, O; Novikova, T; Klementiev, B; Vaudano, E; Berezin, VA; Bock, E. A synthetic NCAM-derived peptide, FGL, protects hippocampal neurons from ischemic insult both in vitro and in vivo. Eur. J. Neurosci 2005, 22, 1589–1596. [Google Scholar]
- Klementiev, B; Novikova, T; Novitskaya, V; Walmod, PS; Dmytriyeva, O; Pakkenberg, B; Berezin, V; Bock, E. A neural cell adhesion molecule-derived peptide reduces neuropathological signs and cognitive impairment induced by Aβ25–35. Neuroscience 2007, 145, 209–224. [Google Scholar]
- Pedersen, MV; Helweg-Larsen, RB; Nielsen, FC; Berezin, V; Bock, E; Penkowa, M. The synthetic NCAM-derived peptide, FGL, modulates the transcriptional response to traumatic brain injury. Neurosci. Lett 2008, 437, 148–153. [Google Scholar]
- Downer, EJ; Cowley, TR; Lyons, A; Mills, KH; Berezin, V; Bock, E; Lynch, MA. A novel anti-inflammatory role of NCAM-derived mimetic peptide, FGL. Neurobiol. Aging 2010, 31, 118–128. [Google Scholar]
- Borcel, E; Pérez-Alvarez, L; Herrero, AI; Brionne, T; Varea, E; Berezin, V; Bock, E; Sandi, C; Venero, C. Chronic stress in adulthood followed by intermittent stress impairs spatial memory and the survival of newborn hippocampal cells in aging animals: Prevention by FGL, a peptide mimetic of neural cell adhesion molecule. Behav. Pharmacol 2008, 19, 41–49. [Google Scholar]
- Popov, VI; Medvedev, NI; Kraev, IV; Gabbott, PL; Davies, HA; Lynch, M; Cowley, TR; Berezin, V; Bock, E; Stewart, MG. A cell adhesion molecule mimetic, FGL peptide, induces alterations in synapse and dendritic spine structure in the dentate gyrus of aged rats: A three-dimensional ultrastructural study. Eur. J. Neurosci 2008, 27, 301–314. [Google Scholar]
- Aonurm-Helm, A; Berezin, V; Bock, E; Zharkovsky, A. NCAM-mimetic, FGL peptide, restores disrupted fibroblast growth factor receptor (FGFR) phosphorylation and FGFR mediated signaling in neural cell adhesion molecule (NCAM)-deficient mice. Brain Res 2010, 1309, 1–8. [Google Scholar]
- Anand, R; Seiberling, M; Kamtchoua, T; Pokorny, R. Tolerability, safety and pharmacokinetics of the FGLL peptide, a novel mimetic of neural cell adhesion molecule, following intranasal administration in healthy volunteers. Clin. Pharmacokinet 2007, 46, 351–358. [Google Scholar]
- Jacobsen, J; Kiselyov, V; Bock, E; Berezin, V. A peptide motif from the second fibronectin module of the neural cell adhesion molecule, NCAM, NLIKQDDGGSPIRHY, is a binding site for the FGF receptor. Neurochem. Res 2008, 33, 2532–2539. [Google Scholar]



| Peptide | Sequence motif localization | In vitro effects | In vivo effects | Refs. |
|---|---|---|---|---|
| Canofin1, 2, and 3 | Interaction sites between FGF2 and FGFR1 | Induction of neurite outgrowth by binding to and activating FGFR. Protection against apoptosis. | N.D. | [50] |
| Hexafin1 and 9 | β6–β7 regions of FGFs | Binding to and activation of FGFR. Protection against apoptosis. | Prolonged retention of social memory in adult rats. | [51,52] |
| Hexafin2 | β6–β7 regions of FGFs | Induction of neurite outgrowth by binding to and activating FGFR. | Prolonged retention of social memory and decreased anxiety-like behavior in adult rats. Alleviation of deficits in activity related to social behavior, including sociability and social novelty, in R6/2 mouse model of Huntington’s disease. | [51,52] |
| Hexafin3, 10, and 17 | β6–β7 regions of FGFs | Induction of neurite outgrowth by binding to and activating FGFR. Protection against apoptosis. | N.D. | [51] |
| Hexafin8 | β6–β7 regions of FGFs | Induction of neurite outgrowth by binding to and activating FGFR. | N.D. | [51] |
| Dekafin1, 2, 3, 5, and 10 | β10–β11 regions of FGFs | Induction of neurite outgrowth by binding to and activating FGFR. | N.D. | [53] |
| Dekafin6, 8, 9, and 17 | β10–β11 regions of FGFs | Induction of neurite outgrowth by binding to and activating FGFR. Protection against apoptosis. | N.D. | [53] |
| DekaCAM | β10–β11 regions of FGFs | Induction of neurite outgrowth by binding to and activating FGFR. | N.D. | [53] |
| FRM | NCAM/FN3(1) | Stimulation of differentiation and neuroprotection. | N.D. | [56] |
| EncaminA | NCAM/FN3(1) | Induction of neurite outgrowth by binding to and activating FGFR and Akt/PKB. | N.D. | [55] |
| EncaminC | NCAM/FN3(1) | Induction of neurite outgrowth by binding to and activating FGFR and Erk. Protection against apoptosis. Enhancement of presynaptic function. | N.D. | [55] |
| EncaminE | NCAM/FN3(1) | Induction of neurite outgrowth by binding to and activating FGFR and Akt/PKB. Protection against apoptosis. Enhancement of presynaptic function. | N.D. | [55] |
| FGL | NCAM/FN3(2) | Induction of neurite outgrowth by binding to and activating FGFR followed by activation of ERK1/2 and Akt/PKB. Activation of FRS2α, ShcA, and PLCγ. Protection against apoptosis. Promotion of synapse formation. Enhancement of presynaptic function. Attenuation of inflammatory impact. | Enhancement of spatial and social memory. Promotion of postnatal sensorimotor development. Protection of hippocampal neurons against ischemic insult. Reduction of neuropathological signs and cognitive impairment in Alzheimer’s disease model. Modulation of the transcriptional response to traumatic brain injury. Attenuation of age-related changes in long-term potentiation and inflammatory signs and prevention of stress-induced dementia. Induction of large changes in the fine structure of dendritic spines in the hippocampus of aged rats related to improved cognitive function. Reversal of depression-like phenotype in NCAM-deficient animals. Amelioration of working memory deficits in rats after neonatal phencyclidine treatment. | [36,45, 54,58–68] |
| BCL | NCAM/FN3(2) | Binding to and activating of FGFR1. Stimulation of neurite outgrowth. | N.D. | [69] |
© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Li, S.; Bock, E.; Berezin, V. Neuritogenic and Neuroprotective Properties of Peptide Agonists of the Fibroblast Growth Factor Receptor. Int. J. Mol. Sci. 2010, 11, 2291-2305. https://doi.org/10.3390/ijms11062291
Li S, Bock E, Berezin V. Neuritogenic and Neuroprotective Properties of Peptide Agonists of the Fibroblast Growth Factor Receptor. International Journal of Molecular Sciences. 2010; 11(6):2291-2305. https://doi.org/10.3390/ijms11062291
Chicago/Turabian StyleLi, Shizhong, Elisabeth Bock, and Vladimir Berezin. 2010. "Neuritogenic and Neuroprotective Properties of Peptide Agonists of the Fibroblast Growth Factor Receptor" International Journal of Molecular Sciences 11, no. 6: 2291-2305. https://doi.org/10.3390/ijms11062291