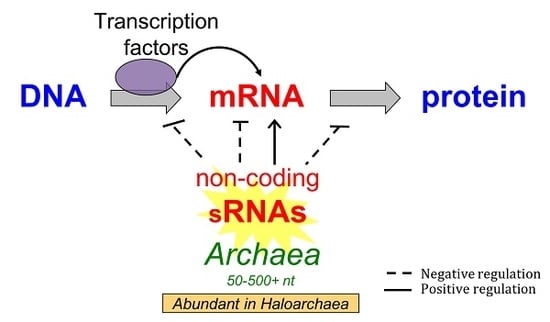
The Non-Coding Regulatory RNA Revolution in Archaea
Abstract
:1. Introduction
2. Identification of sRNAs: What Has Been Found So Far?
3. Best Methods for sRNA Discovery
4. Molecular Regulatory Mechanisms and Targets of Small Non-Coding RNA in Archaea
4.1. Antisense sRNAs
4.2. Intergenic sRNAs
5. Future Prospects
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cech, T.R.; Steitz, J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef] [PubMed]
- Storz, G.; Vogel, J.; Wassarman, K.M. Regulation by small RNAs in Bacteria: expanding frontiers. Mol. Cell 2011, 43, 880–891. [Google Scholar] [CrossRef] [PubMed]
- Wagner, E.G.H.; Romby, P. Chapter Three—Small RNAs in Bacteria and Archaea: Who They Are, What They Do and How They Do It. In Advances in Genetics; Friedmann, T., Dunlap, J.C., Goodwin, S.F., Eds.; Academic Press: Cambridge, MA, USA, 2015; pp. 133–208. [Google Scholar]
- Gelsinger, D.R.; DiRuggiero, J. Transcriptional landscape and regulatory roles of small non-coding RNAs in the oxidative stress response of the haloarchaeon Haloferax volcanii. J. Bacteriol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Prasse, D.; Förstner, K.U.; Jäger, D.; Backofen, R.; Schmitz, R.A. sRNA154 a newly identified regulator of nitrogen fixation in Methanosarcina mazei strain Gö1. RNA Biol. 2017, 14, 1544–1558. [Google Scholar] [CrossRef] [PubMed]
- Fischer, S.; Benz, J.; Späth, B.; Maier, L.K.; Straub, J.; Granzow, M.; Raabe, M.; Urlaub, H.; Hoffmann, J.; Brutschy, B.; et al. The archaeal Lsm protein binds to small RNAs. J. Biol. Chem. 2010, 285, 34429–34438. [Google Scholar] [CrossRef] [PubMed]
- Fischer, S.; Benz, J.; Späth, B.; Jellen-Ritter, A.; Heyer, R.; Dörr, M.; Maier, L.K.; Menzel-Hobeck, C.; Lehr, M.; Jantzer, K.; et al. Regulatory RNAs in Haloferax volcanii. Biochem. Soc. Trans. 2011, 39, 159–162. [Google Scholar] [CrossRef] [PubMed]
- Heyer, R.; Dörr, M.; Jellen-Ritter, A.; Späth, B.; Babski, J.; Jaschinski, K.; Soppa, J.; Marchfelder, A. High throughput sequencing reveals a plethora of small RNAs including tRNA derived fragments in Haloferax volcanii. RNA Biol. 2012, 9, 1011–1018. [Google Scholar] [CrossRef] [PubMed]
- Marchfelder, A.; Fischer, S.; Brendel, J.; Stoll, B.; Maier, L.K.; Jäger, D.; Prasse, D.; Plagens, A.; Schmitz, R.A.; Randau, L. Small RNAs for defence and regulation in Archaea. Extremophiles 2012, 16, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Schmitz-Streit, R.; Jäger, D.; Jellen-Ritter, A.; Babski, J.; Soppa, J.; Marchfelder, A. Archaea employ small RNAs as regulators. In Regulatory RNAs in Prokaryotes; Hess, W.R., Marchfelder, A., Eds.; Springer: Wien, Austria, 2011; pp. 131–145. [Google Scholar]
- Soppa, J.; Straub, J.; Brenneis, M.; Jellen-Ritter, A.; Heyer, R.; Fischer, S.; Granzow, M.; Voss, B.; Hess, W.R.; Tjaden, B.; et al. Small RNAs of the halophilic archaeon Haloferax volcanii. Biochem. Soc. Trans. 2009, 37, 133–136. [Google Scholar] [CrossRef] [PubMed]
- Straub, J.; Brenneis, M.; Jellen-Ritter, A.; Heyer, R.; Soppa, J.; Marchfelder, A. Small RNAs in haloarchaea: Identification, differential expression and biological function. RNA Biol. 2009, 6, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Buddeweg, A.; Sharma, K.; Urlaub, H.; Schmitz, R.A. sRNA41 affects ribosome binding sites within polycistronic mRNAs in Methanosarcina mazei Gö1. Mol. Microbiol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Babski, J.; Haas, K.A.; Näther-Schindler, D.; Pfeiffer, F.; Förstner, K.U.; Hammelmann, M.; Hilker, R.; Becker, A.; Sharma, C.M.; Marchfelder, A.; et al. Genome-wide identification of transcriptional start sites in the haloarchaeon Haloferax volcanii based on differential RNA-Seq (dRNA-Seq). BMC Genom. 2016, 17, 629. [Google Scholar] [CrossRef] [PubMed]
- Morris, K.V.; Mattick, J.S. The rise of regulatory RNA. Nat. Rev. Genet. 2014, 15, 423–437. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, X.; Huang, L.; Guo, H.; Wang, X.J. Potential coexistence of both bacterial and eukaryotic small RNA biogenesis and functional related protein homologs in Archaea. J. Genet. Genom. 2010, 37, 493–503. [Google Scholar] [CrossRef]
- Zander, A.; Willkomm, S.; Ofer, S.; van Wolferen, M.; Egert, L.; Buchmeier, S.; Stöckl, S.; Tinnefeld, P.; Schneider, S.; Klingl, A.; et al. Guide-independent DNA cleavage by archaeal Argonaute from Methanocaldococcus jannaschii. Nat. Microbiol. 2017, 2, 17034. [Google Scholar] [CrossRef] [PubMed]
- Gottesman, S.; Storz, G. Bacterial small RNA regulators: Versatile roles and rapidly evolving variations. Cold Spring Harb Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef] [PubMed]
- Thomason, M.K.; Storz, G. Bacterial antisense RNAs: How many are there and what are they doing? Annu. Rev. Genet. 2010, 44, 167–188. [Google Scholar] [CrossRef] [PubMed]
- Altuvia, S.; Weinstein-Fischer, D.; Zhang, A.; Postow, L.; Storz, G. A small, stable RNA induced by oxidative stress: Role as a pleiotropic regulator and antimutator. Cell 1997, 90, 43–53. [Google Scholar] [CrossRef]
- Smirnov, A.; Förstner, K.U.; Holmqvist, E.; Otto, A.; Günster, R.; Becher, D.; Reinhardt, R.; Vogel, J. Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proc. Natl. Acad. Sci. USA 2016, 113, 11591–11596. [Google Scholar] [CrossRef] [PubMed]
- Wurtzel, O.; Sapra, R.; Chen, F.; Zhu, Y.; Simmons, B.A.; Sorek, R. A single-base resolution map of an archaeal transcriptome. Genome Res. 2010, 20, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Jäger, D.; Sharma, C.M.; Thomsen, J.; Ehlers, C.; Vogel, J.; Schmitz, R.A. Deep sequencing analysis of the Methanosarcina mazei Go1 transcriptome in response to nitrogen availability. Proc. Natl. Acad. Sci. USA 2009, 106, 21878–21882. [Google Scholar] [CrossRef] [PubMed]
- Bernick, D.L.; Dennis, P.P.; Lui, L.M.; Lowe, T.M. Diversity of antisense and other non-coding RNAs in Archaea revealed by comparative small RNA sequencing in four Pyrobaculum species. Front. Microbiol. 2012, 3, 231. [Google Scholar] [CrossRef] [PubMed]
- Toffano-Nioche, C.; Ott, A.; Crozat, E.; Nguyen, A.N.; Zytnicki, M.; Leclerc, F.; Forterre, P.; Bouloc, P.; Gautheret, D. RNA at 92 °C: The non-coding transcriptome of the hyperthermophilic archaeon Pyrococcus abyssi. RNA Biol. 2013, 10, 1211–1220. [Google Scholar] [CrossRef] [PubMed]
- Jäger, D.; Förstner, K.U.; Sharma, C.M.; Santangelo, T.J.; Reeve, J.N. Primary transcriptome map of the hyperthermophilic archaeon Thermococcus kodakarensis. BMC Genom. 2014, 15, 684. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Qi, L.; Guo, Y.; Yue, L.; Li, Y.; Ge, W.; Wu, J.; Shi, W.; Dong, X. Global mapping transcriptional start sites revealed both transcriptional and post-transcriptional regulation of cold adaptation in the methanogenic archaeon Methanolobus psychrophilus. Sci. Rep. 2015, 5, 9209. [Google Scholar] [CrossRef] [PubMed]
- Prasse, D.; Ehlers, C.; Backofen, R.; Schmitz, R.A. Regulatory RNAs in Archaea: First target identification in Methanoarchaea. Biochem. Soc. Trans. 2013, 41, 344–349. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Bachellerie, J.; Rozhdestvensky, T.; Bortolin, M.; Huber, H.; Drungowski, M.; Elge, T.; Brosius, J.; Hüttenhofer, A. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc. Natl. Acad. Sci. USA 2002, 99, 7536–7541. [Google Scholar] [CrossRef] [PubMed]
- Randau, L. RNA processing in the minimal organism Nanoarchaeum equitans. Genome Biology 2012, 13, R63. [Google Scholar] [CrossRef] [PubMed]
- Crits-Christoph, A.; Gelsinger, D.R.; Ma, B.; Wierzchos, J.; Ravel, J.; Davila, A.; Casero, M.C.; DiRuggiero, J. Functional analysis of the Archaea, Bacteria and Viruses from a halite endolithic microbial community. Environ. Microbiol. 2016, 18, 2064–2077. [Google Scholar] [CrossRef] [PubMed]
- Sharma, C.M.; Vogel, J. Differential RNA-seq: The approach behind and the biological insight gained. Curr. Opin. Microbiol. 2014, 19, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, K.; Kish, A.; Pan, M.; Kaur, A.; Reiss, D.J.; King, N.; Hohmann, L.; DiRuggiero, J.; Baliga, N.S. An integrated systems approach for understanding cellular responses to γ radiation. Mol. Syst. Biol. 2006, 2, 47. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, B.S. Small RNAs, big impact: Small RNA pathways in transposon control and their effect on the host stress response. Chromosome Res. 2013, 21, 587–600. [Google Scholar] [CrossRef] [PubMed]
- Capy, P.; Gasperi, G.; Biémont, C.; Bazin, C. Stress and transposable elements: Co-evolution or useful parasites? Heredity 2000, 85, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, D.; Ragunathan, P.T.; Fei, J.; Vanderpool, C.K. A Prophage-encoded small RNA controls metabolism and cell division in Escherichia coli. mSystems 2016, 1, e00021-15. [Google Scholar] [CrossRef] [PubMed]
- Babski, J.; Tjaden, B.; Voss, B.; Jellen-Ritter, A.; Marchfelder, A.; Hess, W.R.; Soppa, J. Bioinformatic prediction and experimental verification of sRNAs in the haloarchaeon Haloferax volcanii. RNA Biol. 2011, 8, 806–816. [Google Scholar] [CrossRef] [PubMed]
- Jaschinski, K.; Babski, J.; Lehr, M.; Burmester, A.; Benz, J.; Heyer, R.; Dörr, M.; Marchfelder, A.; Soppa, J. Generation and phenotyping of a collection of sRNA gene deletion mutants of the Haloarchaeon Haloferax volcanii. PLoS ONE 2014, 9, e90763. [Google Scholar] [CrossRef] [PubMed]
- Jäger, D.; Pernitzsch, S.R.; Richter, A.S.; Backofen, R.; Sharma, C.M.; Schmitz, R.A. An archaeal sRNA targeting cis- and trans-encoded mRNAs via two distinct domains. Nucleic Acids Res. 2012, 40, 10964–10979. [Google Scholar] [CrossRef] [PubMed]
- Märtens, B.; Manoharadas, S.; Hasenöhrl, D.; Manica, A.; Bläsi, U. Antisense regulation by transposon-derived RNAs in the hyperthermophilic archaeon Sulfolobus solfataricus. EMBO Rep. 2013, 14, 527. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.D.; Jackson, S.P. Transcription and translation in Archaea: A mosaic of eukaryal and bacterial features. Trends Microbiol. 1998, 6, 222–228. [Google Scholar] [CrossRef]
- Lalaouna, D.; Prévost, K.; Eyraud, A.; Massé, E. Identification of unknown RNA partners using MAPS. Methods 2017, 117, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Aw, J.G.A.; Shen, Y.; Wilm, A.; Sun, M.; Lim, X.N.; Boon, K.L.; Tapsin, S.; Chan, Y.S.; Tan, C.P.; Sim, A.Y.; Zhang, T. In Vivo Mapping of Eukaryotic RNA Interactomes reveals principles of higher-order organization and regulation. Mol. Cell 2016, 62, 603–617. [Google Scholar] [CrossRef] [PubMed]
- Weidmann, C.A.; Mustoe, A.M.; Weeks, K.M. Direct duplex detection: An emerging tool in the RNA structure analysis toolbox. Trends Biochem. Sci. 2016, 41, 734–736. [Google Scholar] [CrossRef] [PubMed]
- Willkomm, S.; Oellig, C.A.; Zander, A.; Restle, T.; Keegan, R.; Grohmann, D.; Schneider, S. Structural and mechanistic insights into an archaeal DNA-guided Argonaute protein. Nat. Microbiol. 2017, 2, 17035. [Google Scholar] [CrossRef] [PubMed]
- Levy, S.; Portnoy, V.; Admon, J.; Schuster, G. Distinct activities of several RNase J proteins in methanogenic Archaea. RNA Biol. 2011, 8, 1073–1083. [Google Scholar] [CrossRef] [PubMed]
- Gebetsberger, J.; Wyss, L.; Mleczko, A.M.; Reuther, J.; Polacek, N. A tRNA-derived fragment competes with mRNA for ribosome binding and regulates translation during stress. RNA Biol. 2017, 14, 1364–1373. [Google Scholar] [CrossRef] [PubMed]
- Bronsard, J.; Pascreau, G.; Sassi, M.; Mauro, T.; Augagneur, Y.; Felden, B. sRNA and cis-antisense sRNA identification in Staphylococcus aureus highlights an unusual sRNA gene cluster with one encoding a secreted peptide. Sci. Rep. 2017, 7, 4565. [Google Scholar] [CrossRef] [PubMed]
- Bobrovskyy, M.; Frandsen, J.K.; Zhang, J.; Poddar, A.; Azam, M.S.; Henkin, T.M.; Ha, T.; Vanderpool, C.K. Determinants of target prioritization and regulatory hierarchy for the bacterial small RNA SgrS. bioRxiv 2017. [Google Scholar] [CrossRef]




| Number of Genes | Total # sRNAs | # itsRNAs | # asRNAs | # iRNAs | Reference | |
|---|---|---|---|---|---|---|
| Euryarchaeota | ||||||
| Haloferax volcanii | 4023 | 1557 | 77 | 1480 | N/A | [4] |
| Haloferax volcanii | 4023 | 2792 | 395 | 1244 | 1153 | [14] |
| Haloferax volcanii | 4023 | 190 | 145 | 45 | N/A | [8] |
| Methanolobus psychrophilus | 2974 | 2745 | 195 | 1110 | 1440 | [27] |
| Methanosarcina mazei | 3551 | 242 | 199 | 43 | N/A | [23] |
| Thermococcus kodakarensis | 2328 | 1731 | 69 | 1018 | 644 | [26] |
| Pyrococcus abyssi | 1969 | 322 | 107 | 215 | N/A | [25] |
| Archaeoglobus fulgidus | 248 | 45 | 9 | 33 | 3 | [29] |
| Crenarchaeaota | ||||||
| Sulfolobus solfataricus | 3254 | 310 | 125 | 185 | N/A | [22] |
| Pyrobaculum aerophilum, arsenaticum, calidifontis, & islandicum | 2706, 2407, 2200, 2075 | # Not Reported | # Not Reported | 3 | N/A | [24] |
| Nanoarchaeaota | ||||||
| Nanoarchaeum equitans | 553 | # Not Reported | # Not Reported | # Not Reported | N/A | [30] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gelsinger, D.R.; DiRuggiero, J. The Non-Coding Regulatory RNA Revolution in Archaea. Genes 2018, 9, 141. https://doi.org/10.3390/genes9030141
Gelsinger DR, DiRuggiero J. The Non-Coding Regulatory RNA Revolution in Archaea. Genes. 2018; 9(3):141. https://doi.org/10.3390/genes9030141
Chicago/Turabian StyleGelsinger, Diego Rivera, and Jocelyne DiRuggiero. 2018. "The Non-Coding Regulatory RNA Revolution in Archaea" Genes 9, no. 3: 141. https://doi.org/10.3390/genes9030141