Screening for α-Glucosidase-Inhibiting Saponins from Pressurized Hot Water Extracts of Quinoa Husks
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Optimization of the Extraction of Total Saponins
2.2.1. Experimental Design
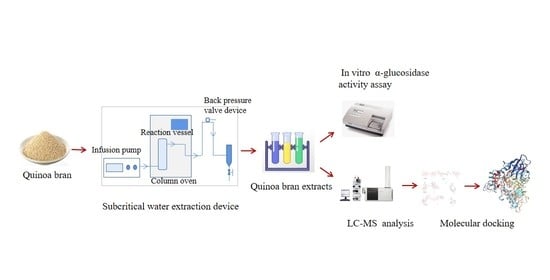
2.2.2. Pressurized Hot Water Extraction of Total Saponins from Quinoa Husks
2.2.3. Determination of the Content of Total Saponins
2.2.4. Optimization of RSM
2.3. LC-MS Analysis of Quinoa Husk Extracts at Optimal Extraction Conditions
2.4. Measurement of α-Glucosidase Activity of Quinoa Husk Extracts at Optimal Extraction Conditions
2.5. Molecular Docking
2.6. Statistical Analysis
3. Results and Discussion
3.1. Optimization of the Extraction Conditions for Total Saponins with RSM
3.2. Identification of Chemical Constituents in Quinoa Husk Extracts
3.3. Inhibitory Effect of Quinoa Husk Extracts on α-Glucosidase
3.3.1. In Vitro Inhibition of α-Glucosidase Activity by Quinoa Husk Extracts
3.3.2. Interaction between α-Glucosidase and Components in Quinoa Husk Extracts
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pereira, E.; Encina-Zelada, C.; Barros, L.; Gonzales-Barron, U.; Cadavez, V.; Ferreira, I.C.F.R. Chemical and nutritional characterization of Chenopodium quinoa Willd (quinoa) grains: A good alternative to nutritious food. Food Chem. 2019, 280, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Melini, V.; Melini, F. Functional components and anti-nutritional factors in gluten-free grains: A focus on quinoa seeds. Foods 2021, 10, 351. [Google Scholar] [CrossRef]
- Viktória, A.; Silva, P.M.; Massuela, D.C.; Khan, M.W.; Hamar, A.; Khajehei, F.; Graeff-Hönninger, S.; Piatti, C. Quinoa (Chenopodium quinoa Willd.): An overview of the potentials of the “golden grain” and socio-economic and environmental aspects of its cultivation and marketization. Foods 2020, 9, 216. [Google Scholar]
- Fan, X.; Guo, H.M.; Teng, C.; Zhang, B.; Blecker, C.; Ren, G.X. Anti-colon cancer activity of novel peptides isolated from in vitro digestion of quinoa protein in Caco-2 cells. Foods 2022, 11, 194. [Google Scholar] [CrossRef]
- Zhao, E.L.; Yang, J. Research progress on extraction and purification technology of saponin from Chenopodium quinoa and its bioactivity. Mol. Plant Breed. 2019, 17, 5816–5821. [Google Scholar]
- Pellegrini, M.; Lucas-Gonzales, R.; Ricci, A.; Fontecha, J.; Fernández-López, J.; Pérez-Álvarez, J.A.; Viuda-Martos, M. Chemical, fatty acid, polyphenolic profile, techno-functional and antioxidant properties of flours obtained from quinoa (Chenopodium quinoa Willd.) seeds. Ind. Crops Prod. 2018, 111, 38–46. [Google Scholar] [CrossRef]
- Ren, Y.P.; Liu, S.X. Effects of separation and purification on structural characteristics of polysaccharide from quinoa (Chenopodium quinoa willd.). Biochem. Biophys. Res. Commun. 2020, 522, 286–291. [Google Scholar] [CrossRef]
- Lin, M.Y.; Han, P.P.; Li, Y.Y.; Wang, W.X. Quinoa secondary metabolites and their biological activities or functions. Molecules 2019, 24, 2512. [Google Scholar] [CrossRef]
- Otterbach, S.; Wellman, G.; Schmckel, S.M. Saponins of quinoa: Structure, function and opportunities. In Quinoa Genome; Springer: Berlin/Heidelberg, Germany, 2021; pp. 119–138. [Google Scholar]
- Hazzam, K.E.; Hafsa, J.; Sobeh, M.; Mhada, M.; Taourirte, M.; Kacimi, K.E.; Yasri, A. An insight into saponins from quinoa (Chenopodium quinoa Willd.): A review. Molecules 2020, 25, 1059. [Google Scholar] [CrossRef]
- Lin, B.J.; Jing, J.J.; Zhang, R.Y.; Du, L.D.; Ji, X.Y.; Ding, K.J.; Xue, P. Research progress of biological activity and chemical structure of saponins (Chenopodium quinoa Willd.). Food Ferment. Ind. 2020, 46, 300–306. [Google Scholar]
- Lim, J.G.; Park, H.; Yoon, K.S. Analysis of saponin composition and comparison of the antioxidant activity of various parts of the quinoa plant (Chenopodium quinoa Willd.). Food Sci. Nutr. 2020, 8, 694–702. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.X.; Yang, X.S.; Zhao, L.; Zhang, F.X.; Hou, Z.H.; Xue, P. Antibacterial activity and mechanism of action saponins from Chenopodium quinoa Willd. husks against foodborne pathogenic bacteria. Ind. Crops Prod. 2020, 149, 112350. [Google Scholar] [CrossRef]
- Zheng, Y.J.; Wang, X.; Zhuang, Y.L.; Li, Y.; Tian, H.L.; Shi, P.Q.; Li, G.F. Isolation of novel ACE-inhibitory and antioxidant peptides from quinoa bran albumin assisted with an in silico approach: Characterization, in vivo antihypertension, and molecular docking. Molecules 2019, 24, 4562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, T.A.; Ke, B.J.; Cheng, C.S.; Wang, J.J.; Wei, B.L.; Lee, C.L. Red quinoa bran extracts protects against carbon tetrachloride-induced liver injury and fibrosis in mice via activation of antioxidative enzyme systems and blocking TGF-β1 pathway. Nutrients 2019, 11, 395. [Google Scholar] [CrossRef] [PubMed]
- Di, T.M.; Yang, S.L.; Du, F.Y.; Zhao, L.; Xia, T.; Zhang, X.F. Cytotoxic and hypoglycemic activity of triterpenoid saponins from Camellia oleifera Abel. seed pomace. Molecules 2017, 22, 1562. [Google Scholar] [CrossRef] [PubMed]
- Nabil, M.; Ghaly, N.S.; Kassem, I.; Grace, M.H.; Melek, F.R. Two triterpenoid saponins with alpha-glucosidase inhibitory activity from Harpullia pendula seed extract. Pharmacogn. J. 2019, 11, 1386–1390. [Google Scholar] [CrossRef]
- Plaza, M.; Marina, M.L. Pressurized hot water extraction of bioactives. Trends Anal. Chem. 2019, 116, 236–247. [Google Scholar] [CrossRef]
- Cheng, Y.; Xue, F.M.; Yu, S.; Du, S.C.; Yang, Y. Subcritical water extraction of natural products. Molecules 2021, 26, 4004. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Zhang, Y.; Taha, A.A.; Ying, Y.; Li, X.P.; Chen, X.Y.; Ma, C. Subcritical water extraction of bioactive components from ginseng roots (Panax ginseng C.A. Mey). Ind. Crops Prod. 2018, 117, 118–127. [Google Scholar] [CrossRef]
- Chi, X.F.; Zhang, G.Y.; Chen, S.L. Subcritical water extraction of sesquiterpene lactones from Inula racemose. Chem. Select. 2020, 5, 488–494. [Google Scholar] [CrossRef]
- Wang, Y.W.; Luan, G.X.; Zhou, W.; Meng, J.; Wang, H.L.; Hu, N.; Suo, Y.R. Subcritical water extraction, UPLC-Triple-TOF/MS analysis and antioxidant activity of anthocyanins from Lycium ruthenicum Murr. Food Chem. 2018, 249, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.J.; Wang, Y.S.; Ma, Y.Y.; Zhu, P.B.; He, J.; Lei, J.D. Optimization of subcritical water extraction of resveratrol from grape seeds by response surface methodology. Appl. Sci. 2017, 7, 321. [Google Scholar] [CrossRef]
- Benmerzoug, A.; Varc-Gaji, J.; Nasti, N.; Guettaf, S. Subcritical water extraction of polyphenols from endemic Algerian plants with medicinal properties. Acta Period. Technol. 2020, 51, 191–206. [Google Scholar] [CrossRef]
- Sakdasri, W.; Arnutpongchai, P.; Phonsavat, S.; Bumrungthaichaichan, E.; Sawangkeaw, R. Pressurized hot water extraction of crude polysaccharides, β-glucan, and phenolic compounds from dried gray oyster mushroom. LWT 2022, 168, 113895. [Google Scholar] [CrossRef]
- Zhang, J.X.; Wen, C.T.; Gu, J.Y.; Ji, C.C.; Duan, Y.Q.; Zhang, H.H. Effects of subcritical water extraction microenvironment on the structure and biological activities of polysaccharides from Lentinus edodes. Int. J. Biol. Macromol. 2018, 123, 1002–1011. [Google Scholar] [CrossRef]
- Melini, V.; Melini, F. Modelling and optimization of ultrasound-assisted extraction of phenolic compounds from black quinoa by response surface methodology. Molecules 2021, 26, 3616. [Google Scholar] [CrossRef]
- Han, Y.M.; Chi, J.W.; Zhang, M.W.; Zhang, R.F.; Fan, S.H.; Huang, F.; Xue, K.M.; Liu, L. Characterization of saponins and phenolic compounds: Antioxidant activity and inhibitory effects on α-glucosidase in different varieties of colored quinoa (Chenopodium quinoa Willd.). Biosci. Biotechnol. Biochem. 2019, 83, 2128–2139. [Google Scholar] [CrossRef]
- Pradeep, P.M.; Sreerama, Y.N. Phenolic antioxidants of foxtail and little millet cultivars and their inhibitory effects on α-amylase and α-glucosidase activities. Food Chem. 2018, 247, 46–55. [Google Scholar] [CrossRef]
- Chen, P.C.; Dlamini, B.S.; Chen, C.R.; Kuo, Y.H.; Shih, W.L.; Lin, Y.S.; Lee, C.H.; Chang, C.I. Structure related α-glucosidase inhibitory activity and molecular docking analyses of phenolic compounds from Paeonia suffruticosa. Med. Chem. Res. 2022, 31, 293–306. [Google Scholar] [CrossRef]
- Auwal, I.M.; Bester, M.J.; Neitz, A.W.; Gaspar, A.R.M. Rational in silico design of novel α-glucosidase inhibitory peptides and in vitro evaluation of promising candidates. Biomed. Pharmacother. 2018, 107, 234–242. [Google Scholar]
- Zhang, H.J.; Li, H.Z.; Zhang, Z.J.; Hou, T.Y. Optimization of ultrasound-assisted extraction of polysaccharides from perilla seed meal by response surface methodology: Characterization and in vitro antioxidant activities. J. Food Sci. 2021, 86, 306–318. [Google Scholar] [CrossRef] [PubMed]
- He, X.F.; Wang, B.; Zhao, B.T.; Yang, F.M. Ultrasonic assisted extraction of quinoa (Chenopodium quinoa Willd.) protein and effect of heat treatment on its in vitro digestion characteristics. Foods 2022, 11, 771. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.X.; Wen, C.T.; Zhang, H.H.; Duan, Y.Q.; Ma, H.L. Recent advances in the extraction of bioactive compounds with subcritical water: A review. Trends Food Sci. Technol. 2020, 95, 183–195. [Google Scholar] [CrossRef]
- Dong, Q.; Tan, L.; Hu, N.; Wang, H.L. Study on the optimization of the extraction technology by response surface methodology and inhibitory activity of α-glucoside of total saponins from the bran of Chenopodium quinoa Willd. Nat. Prod. Res. Dev. 2021, 33, 935–942. [Google Scholar]
- Yang, J.; Gao, F.X.; Yang, M.; Bian, H.Y.; Zhang, S.G.; Zhao, E.L. Study on microwave-assisted extraction and antioxidant activity of total saponins from Chenopodium quinoa peel. Food. Mach. 2017, 12, 148–153. [Google Scholar]
- Yang, D. Optimization of supercritical CO2 extraction process of quinoa bran saponin. Food Res. Dev. 2019, 40, 149–154. [Google Scholar]
- Xu, X.Q.; Zhao, W.T.; Miao, L.X.; Huo, N.R. Extraction and purification of total saponins from quinoa bran. Sci. Technol. Food Ind. 2017, 38, 215–220. [Google Scholar]
- Dong, Q.; Hu, N.; Yue, H.L.; Wang, H.L.; Ku, J.L. Identification of α -glucosidase inhibitors from the bran of Chenopodium quinoa Willd. by surface plasmon resonance coupled with ultra-performance liquid chromatography and quadrupole-time-of-flight-mass spectrometry. J. Chromatogr. B 2021, 1181, 122919. [Google Scholar] [CrossRef]
- Jiang, S.R.; Zhao, X.H.; Liu, C.; Dong, Q.; Mei, L.J.; Chen, C.; Shao, Y.; Tao, Y.D.; Yue, H.L. Identification of phenolic compounds in fruits of Ribes stenocarpum Maxim. by UHPLC-QTOF/MS and their hypoglycemic effects in vitro and in vivo. Food Chem. 2020, 344, 128568. [Google Scholar] [CrossRef]
- Tang, Y.; Zhang, B.; Li, X.H.; Chen, P.X.; Zhang, H.; Liu, R.H.; Tsao, R. Bound phenolics of quinoa seeds released by acid, alkaline, and enzymatic treatments and their antioxidant and α-glucosidase and pancreatic lipase inhibitory effects. J. Agric. Food Chem. 2016, 64, 1712–1719. [Google Scholar] [CrossRef]
- Madl, T.; Sterk, H.; Mittelbach, M.; Rechberger, G.N. Tandem mass spectrometric analysis of a complex triterpene saponin mixture of Chenopodium quinoa. J. Am. Soc. Mass Spectrom. 2006, 17, 795–806. [Google Scholar] [PubMed]
- Xiong, H.; Ding, X.; Yang, G.Z.; Yang, G.Z.; Mei, Z.N. Triterpene saponins from the stems of Entada phaseoloides. Planta Med. 2014, 80, 710–718. [Google Scholar] [CrossRef] [PubMed]
- Kuljanabhagavad, T.; Wink, M. Biological activities and chemistry of saponins from Chenopodium quinoa Willd. Phytochem. Rev. 2009, 8, 473–490. [Google Scholar] [CrossRef]
- Yuan, T.; Guo, X.F.; Shao, S.Y.; An, R.M. Characterization and identification of flavonoids from Bambusa chungii leaves extract by UPLC-ESI-Q-TOF-MS/MS. Acta Chromatogr. 2020, 33, 281–294. [Google Scholar] [CrossRef]
- Aghakhani, F.; Kharazin, N.; Gooini, Z.L. Flavonoid constituents of phlomis (Lamiaceae) species using liquid chromatography mass spectrometry. Phytochem. Anal. 2017, 29, 180–195. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Kumar, S.; Bajpai, V.; Reddy, T.J.; Rameshkumar, K.B.; Kumaret, B. Structural characterization of flavonoid C- and O-glycosides in an extract of Adhatoda vasica leaves by liquid chromatography with quadrupole time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2015, 29, 1095–1106. [Google Scholar] [CrossRef]
- Lazo-Vélez, M.A.; Guajardo-Flores, D.; Mata-Ramírez, D.; Gutiérrez-Uribe, J.A.; Serna-Saldivar, S.O. Characterization and quantitation of triterpenoid saponins in raw and sprouted Chenopodium berlandieri spp. (Huauzontle) grains subjected to germination with or without selenium stress conditions. J. Food Sci. 2016, 19, 19–26. [Google Scholar] [CrossRef]
- Xu, Y.; Rashwan, A.K.; Ge, Z.W.; Li, Y.T.; Ge, H.J.; Li, J.X.; Xie, J.H.; Liu, S.Y.; Fang, J.; Cheng, K.J.; et al. Identification of a novel α-glucosidase inhibitor from Melastoma dodecandrum Lour. fruits and its effect on regulating postprandial blood glucose. Food Chem. 2022, 399, 133999. [Google Scholar] [CrossRef]
- Fu, M.H.; Shen, W.X.; Gao, W.Z.; Namujia, L.X.; Yang, X.; Cao, J.W.; Sun, L.J. Essential moieties of myricetins, quercetins and catechins for binding and inhibitory activity against α-Glucosidase. Bioorg. Chem. 2021, 115, 105235. [Google Scholar] [CrossRef]
- Nguyen, T.P.; Le, T.D.; Phan, N.M.; Bui, T.D.; Mai, D.T. Triterpene saponins with α-glucosidase inhibition and cytotoxic activity from the leaves of Schefflera sessiliflora. J. Asian Nat. Prod. Res. 2015, 18, 542–549. [Google Scholar] [CrossRef]
- Şöhretoğlu, D.; Sari, S.; Barut, B.; Özel, A. Discovery of potent α-glucosidase inhibitor flavonols: Insights into mechanism of action through inhibition kinetics and docking simulations. Bioorg. Chem. 2018, 79, 257–264. [Google Scholar] [CrossRef]
- Fan, X.Z.; Zhu, Y.L.; Yuan, R.W.; Deng, L.; Hou, C.; Li, W.; Liu, T.; Kong, X.Q.; Zhang, L.J.; Liao, H.B. Terpenoids with α-glucosidase inhibitory activity from Rhododendron minutiflorum Hu. Phytochemistry 2022, 196, 113083. [Google Scholar] [CrossRef] [PubMed]
- Miller, N.; Joubert, E. Critical assessment of in vitro screening of α-glucosidase inhibitors from plants with acarbose as a reference standard. Planta Med. 2022, 88, 1078–1091. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.L.; Kong, Y.C.; Miao, J.Y.; Mei, X.Y.; Cao, X.Y. Spectroscopy and molecular docking analysis reveal structural specificity of flavonoids in the inhibition of α-glucosidase activity. Int. J. Biol. Macromol. 2019, 152, 981–989. [Google Scholar] [CrossRef]
- Luo, J.G.; Ma, L.; Kong, L.Y. New triterpenoid saponins with strong α-glucosidase inhibitory activity from the roots of Gypsophila oldhamiana. Bioorg. Med. Chem. 2008, 16, 2912–2920. [Google Scholar] [CrossRef] [PubMed]
- Li, S.Y.; Yin, L.; Yi, J.Z.; Zhang, L.M.; Yang, L.Q. Insight into interaction mechanism between theaflavin-3-gallate and α-glucosidase using spectroscopy and molecular docking analysis. J. Food Biochem. 2021, 45, e13500. [Google Scholar] [CrossRef] [PubMed]
- Proença, C.; Freitas, M.; Ribeiro, D.; Oliveira, E.F.T.; Sousa, J.L.C.; Tomé, S.M. α-Glucosidase inhibition by flavonoids: An in vitro and in silico structure-activity relationship study. J. Enzym. Inhib. Med. Chem. 2017, 32, 1216–1228. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.F.; Hu, X.; Xu, X.M.; Zhang, G.W.; Gong, D.M. Inhibitory mechanism of two allosteric inhibitors, oleanolic acid and ursolic acid on α-glucosidase. Int. J. Biol. Macromol. 2018, 107, 1844–1855. [Google Scholar] [CrossRef]
- Dou, F.; Xi, M.M.; Wang, J.X.; Tian, X.R.; Wen, A.D. α Glucosidase and α amylase inhibitory activities of saponins from traditional Chinese medicines in the treatment of diabetes mellitus. Die Pharm. Int. J. Pharm. Sci. 2013, 68, 300–304. [Google Scholar]
- Murugesu, S.; Ibrahim, Z.; Ahmed, Q.U.; Yusoff, N.I.N.; Uzir, B.F.; Perumal, V.; Abas, F.; Saari, K.; El-Seedi, H.; Khatib, A. Characterization of α-Glucosidase Inhibitors from Clinacanthus nutans lindau leaves by gas chromatography-mass Spectrometry-based metabolomics and molecular docking simulation. Molecules 2018, 23, 2402. [Google Scholar] [CrossRef] [Green Version]







| Factors | Symbols | Levels | ||
|---|---|---|---|---|
| −1 | 0 | 1 | ||
| Extraction Flow Rate (mL/min) | A | 2 | 4 | 6 |
| Extraction Temperature (°C) | B | 150 | 180 | 210 |
| Extraction Time (min) | C | 30 | 45 | 60 |
| Run Order | Extraction Flow Rate (mL/min) A | Extraction Temperature (°C) B | Extraction Time (min) C | The Yield of Total Saponins (mg/g) Y |
|---|---|---|---|---|
| 1 | 2 | 180 | 60 | 16.77 ± 0.56 |
| 2 | 4 | 180 | 45 | 18.67 ± 1.47 |
| 3 | 2 | 150 | 45 | 19.16± 0.38 |
| 4 | 4 | 210 | 60 | 21.89 ± 1.81 |
| 5 | 4 | 180 | 45 | 19.61 ± 1.05 |
| 6 | 6 | 180 | 30 | 17.02 ± 0.30 |
| 7 | 6 | 180 | 60 | 18.81 ± 0.18 |
| 8 | 6 | 150 | 45 | 22.54 ± 0.67 |
| 9 | 6 | 210 | 45 | 22.90 ± 1.29 |
| 10 | 4 | 180 | 45 | 19.01 ± 0.75 |
| 11 | 2 | 210 | 45 | 23.57 ± 0.69 |
| 12 | 2 | 180 | 30 | 17.88 ± 0.83 |
| 13 | 4 | 180 | 45 | 19.31 ± 1.28 |
| 14 | 4 | 150 | 30 | 19.74 ± 0.64 |
| 15 | 4 | 150 | 60 | 18.01 ± 0.29 |
| 16 | 4 | 180 | 45 | 18.91 ± 1.53 |
| 17 | 4 | 210 | 30 | 20.34 ± 1.58 |
| Extraction Technique | Solvent Used | Extraction Temperature (°C) | Extraction Time (min) | Solid/Solvent | Other Parameters | The Yield of Total Saponins (mg/g) | Ref |
|---|---|---|---|---|---|---|---|
| Ultrasonic-assisted extraction | 75% EtOH | 45 | 90 | 1:15 | - | 23.7 | [35] |
| Microwave-assisted extraction | 68% EtOH | - | 10 | 1:32 | Power 455 W | 26.32 | [35] |
| Supercritical CO2 extraction | 74% EtOH | 60 | 96 | - | Pressure 37 MPa | 9.6 | [37] |
| Solvent reflux extraction | 72% EtOH | 72 | 147 | 1:20.8 | - | 16.85 | [38] |
| Pressurized hot water extraction | Water | 210 | 50 | - | Flow rate 2 mL/min | 23.06 |
| NO. | RT (min) | [M-H] (m/z) | MS/MS Fragments | Formula | Compound | Ref |
|---|---|---|---|---|---|---|
| 1 | 8.04 | 755.2144 | 755.2144, 300.0274. | C33H40O20 | Quercetin 3-O-(2,6-di-α-l-rhamnopyranosyl) -β-d-galactopyranoside | [39] |
| 2 | 8.77 | 739.2213 | 739.2213, 285.0417. | C33H40O19 | kaempferol 3-O-(2,6-di-α-l-rhamnopyranosyl) -β-d-galactopyranoside | [39] |
| 3 | 9.95 | 477.0687 | 477.0687, 301.0370. | C21H18O13 | quercetin 3-O-β-d-glucuronopyranoside | [39] |
| 4 | 10.11 | 479.3041 | 479.3041, 319.1914, 159.1016. | C21H20O13 | Myricetin-3-O-β-d-galactopyranoside | [40] |
| 5 | 11.30 | 187.0096 | 187.0096, 123.0821, 97.0676 | C9H8O3 | p-Coumaric acid | [41] |
| 6 | 12.76 | 957.4882 | 957.4882, 795.4309, 633.3719, 501.3251, | C48H76O19 | Serjanic acid 3-O-[β-d-glucopyranosyl-(1-3)-α-l- arabinopyranosyl]-28-O-β-d-glucopyranoside | [42] |
| 7 | 13.36 | 827.4482 [M+COOH]− | 827.4482, 781.4515, 619.3924, 487.3436 | C41H66O14 | 3β,15α,16α-trihydroxy-18β-olean-12-en-28-oic acid 28-O-α-l-arabinopyanosyl-(1-3)-β-d- glucopyranosyl ester | [43] |
| 8 | 14.05 | 301.0366 | 301.0366, 151.0033 | C15H10O7 | Quercetin | [41] |
| 9 | 15.34 | 1017.4968 [M+COOH]− | 855.4428, 809.4467, 647.3884, 515.3427 | C48H76O20 | 3-O-β-d-glucopyranosyl-(1-3)-O-α-l-arabinopyranosyl phytolaccagenic acid 28-O-β-d- glucopyranosyl ester | - |
| 10 | 16.42 | 855.4429 [M+COOH]− | 855.4429, 809.4522, 647.3887, 515.3412, | C42H66O15 | O-β-d-glucopyranosyl-(1-3)-O-α-l- arabinopyranosyl phytolaccagenic acid | [39] |
| 11 | 17.10 | 809.4465 | 809.4465, 647.3887, 471.3520 | C42H66O15 | 3-O-β-d-glucuronopyranosyl hederagenin 28-O-β-d-glucopyranosyl ester | [42] |
| 12 | 17.63 | 973.5057 [M+COOH]− | 973.5057, 765.4548, 603.3971, 471.3520 | C47H76O18 | Hederagenin 3-O-[β-d-glucopyranosyl-(1,3)-α- l-arabinopyranosyl]-28-O-β-d-glucopyranoside | [39] |
| 13 | 18.49 | 969.4519 | 969.4519, 925.4610, 809.4471, 471.3521 | C47H70O21 | basellasaponin A | [39] |
| 14 | 19.41 | 793.4496 | 793.4496, 631.3915, 455.3551 | C42H66O14 | 3-O-β-d-glucuronopyranosyl oleanolic acid 28-O-β-d-glucopyranosyl ester | [42] |
| 15 | 19.85 | 693.3514 | 693.3514, 647.3458, 515.3437 | C36H56O10 | 3-O-α-l-arabinopyranosyl phytolaccagenic acid | [39] |
| 16 | 20.51 | 953.4559 | 953.4559, 909.4650, 793.450, 631.3938, 455.3565 | C48H76O19 | O-β-d-glucopyranosyl-(1-3)-β-d- glucuronopyranosyl oleanolic acid 28-O-β-d-glucopyranosyl ester | - |
| 17 | 20.99 | 851.4580 | 851.4580, 807.3955, 691.3794, 515.3426 | C43H66O17 | 3-O-β-d-glucopyranosyl-(1-3)-β-d glucuronopyranosyl phytolaccagenic acid | - |
| 18 | 22.04 | 647.3820 | 647.3820, 471.3506 | C36H56O10 | Hederagenin 3-O-β-d-glucuronopyranoside | - |
| 19 | 25.32 | 631.3916 | 631.3916, 455.3558 | C36H56O9 | 3-O-β-d-glucuronopyranosyl oleanolic acid | [44] |
| 20 | 26.65 | 779.3988 | 779.3988, 647.3878, 471.3512 | C41H64O14 | Hederagenin 3-O-β-d-xylopyranosyl-(1-3)-β-d glucuronopyranoside | - |
| 21 | 27.51 | 791.3892 | 791.3892, 631.3940, 455.3574 | C42H66O14 | 14 isomer | [42] |
| 22 | 29.97 | 763.4042 | 763.4042, 631.3930, 455.3562 | C41H64O13 | oleanolic acid 3-O-β-d-xylopyranosyl-(1-3)-β-d glucuronopyranoside | [44] |
| 23 | 30.79 | 763.4042 | 763.4042, 631.3930, 455.3562 | C41H64O13 | 22 isomer | [44] |
| Flavonoids | Triterpenoid Saponins | Acarbose | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Compound Number | 1 | 2 | 3 | 8 | 9 | 14 | 16 | 19 | |
| Affinity (kcal/mol) | −8.6 | −8.1 | −9.7 | −8.9 | −11.6 | −12.6 | −12.2 | −12.7 | −8.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, R.; Li, J.; Hu, N.; Wang, H.; Cao, J.; Chi, X.; Dong, Q. Screening for α-Glucosidase-Inhibiting Saponins from Pressurized Hot Water Extracts of Quinoa Husks. Foods 2022, 11, 3026. https://doi.org/10.3390/foods11193026
Su R, Li J, Hu N, Wang H, Cao J, Chi X, Dong Q. Screening for α-Glucosidase-Inhibiting Saponins from Pressurized Hot Water Extracts of Quinoa Husks. Foods. 2022; 11(19):3026. https://doi.org/10.3390/foods11193026
Chicago/Turabian StyleSu, Rong, Jing Li, Na Hu, Honglun Wang, Jingya Cao, Xiaofeng Chi, and Qi Dong. 2022. "Screening for α-Glucosidase-Inhibiting Saponins from Pressurized Hot Water Extracts of Quinoa Husks" Foods 11, no. 19: 3026. https://doi.org/10.3390/foods11193026