Geometry-Based Deep Learning in the Natural Sciences
Definition
:1. Background
1.1. Historical Perspective on Geometry
1.2. The Explanatory Power of Geometry
2. Geometrical Explanations of Adaptive Immunity
2.1. Overview
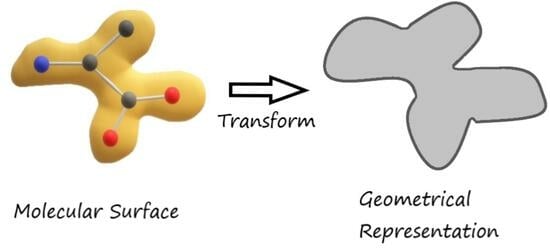
2.2. The Geometry of Molecular Interactions
2.3. Deep Learning and Geometrical Modeling
2.4. Perspectives on Deep Learning
3. Modeling the Molecular Surfaces of Immunity
4. Modeling the Molecular Surface of Proteins
5. Geometrical Explanations of Cognition
6. Abstractive Models of Complex Systems
7. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- The Stanford Encyclopedia of Philosophy; Stanford University: Stanford, CA, USA, 2019; Available online: https://plato.stanford.edu/entries/pythagoreanism (accessed on 25 April 2023).
- Proust, D. The Harmony of the Spheres from Pythagoras to Voyager. Proc. Int. Astron. Union 2009, 5, 358–367. [Google Scholar] [CrossRef] [Green Version]
- Murschel, A. The Structure and Function of Ptolemy’s Physical Hypotheses of Planetary Motion. J. Hist. Astron. 1995, 26, 33–61. [Google Scholar] [CrossRef]
- Van Helden, A. Galileo, telescopic astronomy, and the Copernican system. In Planetary Astronomy from the Renaissance to the Rise of Astrophysics; Taton, R., Wilson, C., Eds.; Cambridge University Press: Cambridge, UK, 1989. [Google Scholar]
- Settle, T.B. An Experiment in the History of Science: With a simple but ingenious device Galileo could obtain relatively precise time measurements. Science 1961, 133, 19–23. [Google Scholar] [CrossRef]
- Truesdell, C. History of Classical Mechanics (Part I). Naturwissenschaften 1976, 63, 53–62. [Google Scholar] [CrossRef]
- Kleppner, D. A short history of atomic physics in the twentieth century. Rev. Mod. Phys. 1999, 71, S78. [Google Scholar] [CrossRef]
- Hartshorne, R. Geometry: Euclid and Beyond; Springer Science & Business Media: New York, NY, USA, 2013. [Google Scholar]
- Arthur, R.T. Minkowski Spacetime and the Dimensions of the Present. Philos. Found. Phys. 2006, 1, 129–155. [Google Scholar]
- Friedman, R. Themes of advanced information processing in the primate brain. AIMS Neurosci. 2020, 7, 373. [Google Scholar] [CrossRef] [PubMed]
- Waddell, W.W. The Parmenides of Plato; James Maclehose and Sons: Glasgow, UK, 1894. [Google Scholar]
- Barrett, T.W. Conservation of information. Acta Acust. United Acust. 1972, 27, 44–47. [Google Scholar]
- Friedman, R. Detecting Square Grid Structure in an Animal Neuronal Network. NeuroSci 2022, 3, 91–103. [Google Scholar] [CrossRef]
- Linial, N.; London, E.; Rabinovich, Y. The geometry of graphs and some of its algorithmic applications. Combinatorica 1995, 15, 215–245. [Google Scholar] [CrossRef]
- Goswami, S.; Murthy, C.A.; Das, A.K. Sparsity measure of a network graph: Gini index. Inf. Sci. 2018, 462, 16–39. [Google Scholar] [CrossRef] [Green Version]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention is all you need. In Proceedings of the 31st Conference on Neural Information Processing System, Long Beach, CA, USA, 4–9 December 2017; pp. 5998–6008. [Google Scholar]
- Chen, L.; Lu, K.; Rajeswaran, A.; Lee, K.; Grover, A.; Laskin, M.; Abbeel, P.; Srinivas, A.; Mordatch, I. Decision Transformer: Reinforcement Learning via Sequence Modeling. Adv. Neural Inf. Process. Syst. 2021, 34, 15084–15097. [Google Scholar]
- Odum, E.P. Energy flow in ecosystems—A historical review. Am. Zool. 1968, 8, 11–18. [Google Scholar] [CrossRef]
- Hu, Y.; Buehler, M.J. Deep language models for interpretative and predictive materials science. APL Mach. Learn. 2023, 1, 010901. [Google Scholar] [CrossRef]
- Schmidhuber, J. Learning to Control Fast-Weight Memories: An Alternative to Dynamic Recurrent Networks. Neural Comput. 1992, 4, 131–139. [Google Scholar] [CrossRef]
- Omotehinwa, T.O.; Ramon, S.O. Fibonacci Numbers and Golden Ratio in Mathematics and Science. Int. J. Comput. Inf. Technol. 2013, 2, 630–638. [Google Scholar]
- Kelley, R.; Ideker, T. Systematic interpretation of genetic interactions using protein networks. Nat. Biotechnol. 2005, 23, 561–566. [Google Scholar] [CrossRef] [Green Version]
- Boone, C.; Bussey, H.; Andrews, B.J. Exploring genetic interactions and networks with yeast. Nat. Rev. Genet. 2007, 8, 437–449. [Google Scholar] [CrossRef] [PubMed]
- Peng, R. The reproducibility crisis in science: A statistical counterattack. Significance 2015, 12, 30–32. [Google Scholar] [CrossRef] [Green Version]
- Gardenier, J.; Resnik, D. The Misuse of Statistics: Concepts, Tools, and a Research Agenda. Account. Res. Policies Qual. Assur. 2002, 9, 65–74. [Google Scholar] [CrossRef]
- Davis, M.M.; Bjorkman, P.J. T-cell antigen receptor genes and T-cell recognition. Nature 1988, 334, 395–402. [Google Scholar] [CrossRef]
- Germain, R.N. MHC-dependent antigen processing and peptide presentation: Providing ligands for T lymphocyte activation. Cell 1994, 76, 287–299. [Google Scholar] [CrossRef] [PubMed]
- Garstka, M.A.; Fish, A.; Celie, P.H.; Joosten, R.P.; Janssen, G.M.; Berlin, I.; Hoppes, R.; Stadnik, M.; Janssen, L.; Ovaa, H.; et al. The first step of peptide selection in antigen presentation by MHC class I molecules. Proc. Natl. Acad. Sci. USA 2015, 112, 1505–1510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedman, R. A Hierarchy of Interactions between Pathogenic Virus and Vertebrate Host. Symmetry 2022, 14, 2274. [Google Scholar] [CrossRef]
- Wong, F.; Krishnan, A.; Zheng, E.J.; Stark, H.; Manson, A.L.; Earl, A.M.; Jaakkola, T.; Collins, J.J. Benchmarking AlphaFold-enabled molecular docking predictions for antibiotic discovery. Mol. Syst. Biol. 2022, 18, e11081. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, K.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Lythe, G.; Callard, R.E.; Hoare, R.L.; Molina-Paris, C. How many TCR clonotypes does a body maintain? J. Theor. Biol. 2016, 389, 214–224. [Google Scholar] [CrossRef] [Green Version]
- Hie, B.; Zhong, E.D.; Berger, B.; Bryson, B. Learning the language of viral evolution and escape. Science 2021, 371, 284–288. [Google Scholar] [CrossRef]
- Krangel, M.S. Mechanics of T cell receptor gene rearrangement. Curr. Opin. Immunol. 2009, 21, 133–139. [Google Scholar] [CrossRef] [Green Version]
- Schuldt, N.J.; Binstadt, B.A. Dual TCR T Cells: Identity Crisis or Multitaskers? J. Immunol. 2019, 202, 637–644. [Google Scholar] [CrossRef] [Green Version]
- Hodgkin, P.D. Modifying clonal selection theory with a probabilistic cell. Immunol. Rev. 2018, 285, 249–262. [Google Scholar] [CrossRef] [Green Version]
- Alt, F.W.; Oltz, E.M.; Young, F.; Gorman, J.; Taccioli, G.; Chen, J. VDJ recombination. Immunol. Today 1992, 13, 306–314. [Google Scholar] [CrossRef]
- Friedman, R. A Perspective on Information Optimality in a Neural Circuit and Other Biological Systems. Signals 2022, 3, 410–427. [Google Scholar] [CrossRef]
- Rooklin, D.; Wang, C.; Katigbak, J.; Arora, P.S.; Zhang, Y. AlphaSpace: Fragment-Centric Topographical Mapping To Target Protein–Protein Interaction Interfaces. J. Chem. Inf. Model. 2015, 55, 1585–1599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCafferty, C.L.; Marcotte, E.M.; Taylor, D.W. Simplified geometric representations of protein structures identify complementary interaction interfaces. Proteins: Struct. Funct. Bioinform. 2021, 89, 348–360. [Google Scholar] [CrossRef] [PubMed]
- Dai, B.; Bailey-Kellogg, C. Protein interaction interface region prediction by geometric deep learning. Bioinformatics 2021, 37, 2580–2588. [Google Scholar] [CrossRef]
- Montemurro, A.; Schuster, V.; Povlsen, H.R.; Bentzen, A.K.; Jurtz, V.; Chronister, W.D.; Crinklaw, A.; Hadrup, S.R.; Winther, O.; Peters, B.; et al. NetTCR-2.0 enables accurate prediction of TCR-peptide binding by using paired TCRα and β sequence data. Commun. Biol. 2021, 4, 1060. [Google Scholar] [CrossRef]
- Bradley, P. Structure-based prediction of T cell receptor: Peptide-MHC interactions. ELife 2023, 12, e82813. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.; Verkuil, R.; Liu, J.; Lin, Z.; Hie, B.; Sercu, T.; Lerer, A.; Rives, A. Learning inverse folding from millions of predicted structures. In Proceedings of the 39th International Conference on Machine Learning, Proceedings of Machine Learning Research, Baltimore, MD, USA, 17–23 July 2022; pp. 8946–8970. [Google Scholar]
- Taylor, R.; Kardas, M.; Cucurull, G.; Scialom, T.; Hartshorn, A.; Saravia, E.; Poulton, A.; Kerkez, V.; Stojnic, R. Galactica: A Large Language Model for Science. arXiv 2022, arXiv:2211.09085. [Google Scholar]
- Grezes, F.; Allen, T.; Blanco-Cuaresma, S.; Accomazzi, A.; Kurtz, M.J.; Shapurian, G.; Henneken, E.; Grant, C.S.; Thompson, D.M.; Hostetler, T.W.; et al. Improving astroBERT using Semantic Textual Similarity. arXiv 2022, arXiv:2212.00744. [Google Scholar]
- Stirling, A. Disciplinary dilemma: Working across research silos is harder than it looks. Guardian 2014. Available online: https://www.theguardian.com/science/political-science/2014/jun/11/science-policy-research-silos-interdisciplinarity (accessed on 10 May 2023).
- Opik, E.J. About dogma in science, and other recollections of an astronomer. Annu. Rev. Astron. Astrophys. 1977, 15, 1–18. [Google Scholar] [CrossRef]
- Neyman, J. Frequentist Probability and Frequentist Statistics. Synthese 1977, 36, 97–131. [Google Scholar] [CrossRef]
- Quinn, J.F.; Dunham, A.E. On Hypothesis Testing in Ecology and Evolution. Am. Nat. 1983, 122, 602–617. [Google Scholar] [CrossRef]
- Lin, Z.; Akin, H.; Rao, R.; Hie, B.; Zhu, Z.; Lu, W.; dos Santos Costa, A.; Fazel-Zarandi, M.; Sercu, T.; Candido, S.; et al. Language models of protein sequences at the scale of evolution enable accurate structure prediction. BioRxiv 2022. bioRxiv:2022.07.20.500902. [Google Scholar]
- Brants, T.; Popat, A.C.; Xu, P.; Och, F.J.; Dean, J. Large language models in machine translation. In Proceedings of the 2007 Joint Conference on Empirical Methods in Natural Language Processing and Computational Natural Language Learning (EMNLP-CoNLL), Prague, Czech Republic, 28–30 June 2007; pp. 858–867. [Google Scholar]
- Creswell, A.; Shanahan, M.; Higgins, I. Selection-inference: Exploiting large language models for interpretable logical reasoning. arXiv 2022, arXiv:2205.09712. [Google Scholar]
- Brown, T.; Mann, B.; Ryder, N.; Subbiah, M.; Kaplan, J.D.; Dhariwal, P.; Neelakantan, A.; Shyam, P.; Sastry, G.; Askell, A.; et al. Language Models are Few-Shot Learners. Adv. Neural Inf. Process. Syst. 2020, 33, 1877–1901. [Google Scholar]
- Cappuccio, A.; Jensen, S.T.; Hartmann, B.M.; Sealfon, S.C.; Soumelis, V.; Zaslavsky, E. Deciphering the combinatorial landscape of immunity. Elife 2020, 9, e62148. [Google Scholar] [CrossRef]
- Hou, X.L.; Wang, L.; Ding, Y.L.; Xie, Q.; Diao, H.Y. Current status and recent advances of next generation sequencing techniques in immunological repertoire. Genes Immun. 2016, 17, 153–164. [Google Scholar] [CrossRef]
- Lutz, I.D.; Wang, S.; Norn, C.; Courbet, A.; Borst, A.J.; Zhao, Y.T.; Dosey, A.; Cao, L.; Xu, J.; Leaf, E.M.; et al. Top-down design of protein architectures with reinforcement learning. Science 2023, 380, 266–273. [Google Scholar] [CrossRef]
- Silver, D.; Huang, A.; Maddison, C.J.; Guez, A.; Sifre, L.; Van Den Driessche, G.; Schrittwieser, J.; Antonoglou, I.; Panneershelvam, V.; Lanctot, M.; et al. Mastering the game of Go with deep neural networks and tree search. Nature 2016, 529, 484–489. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R. Tokenization in the Theory of Knowledge. Encyclopedia 2023, 3, 380–386. [Google Scholar] [CrossRef]
- Puy, A.; Beneventano, P.; Levin, S.A.; Lo Piano, S.; Portaluri, T.; Saltelli, A. Models with higher effective dimensions tend to produce more uncertain estimates. Sci. Adv. 2022, 8, eabn9450. [Google Scholar] [CrossRef]
- Pan, Y.; Wang, L.; Feng, Z.; Xu, H.; Li, F.; Shen, Y.; Zhang, D.; Liu, W.J.; Gao, G.F.; Wang, Q. Characterisation of SARS-CoV-2 variants in Beijing during 2022: An epidemiological and phylogenetic analysis. Lancet 2023, 401, 664–672. [Google Scholar] [CrossRef]
- Govindarajan, S.; Recabarren, R.; Goldstein, R.A. Estimating the total number of protein folds. Proteins Struct. Funct. Bioinform. 1999, 35, 408–414. [Google Scholar] [CrossRef]
- Young, L.; Jernigan, R.L.; Covell, D.G. A role for surface hydrophobicity in protein-protein recognition. Protein Sci. 1994, 3, 717–729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, S.; Thornton, J.M. Analysis of protein-protein interaction sites using surface patches. J. Mol. Biol. 1997, 272, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Ma, B.; Wolfson, H.; Nussinov, R. Conservation of polar residues as hot spots at protein interfaces. Proteins Struct. Funct. Bioinform. 2000, 39, 331–342. [Google Scholar] [CrossRef]
- Richards, F.M. 1977. Areas, Volumes, Packing, and Protein Structure. Annu. Rev. Biophys. Bioeng. 1977, 6, 151–176. [Google Scholar] [CrossRef]
- Bajaj, C.L.; Pascucci, V.; Shamir, A.; Holt, R.J.; Netravali, A.N. Dynamic maintenance and visualization of molecular surfaces. Discret. Appl. Math. 2003, 127, 23–51. [Google Scholar] [CrossRef] [Green Version]
- Gainza, P.; Sverrisson, F.; Monti, F.; Rodola, E.; Boscaini, D.; Bronstein, M.M.; Correia, B.E. Deciphering interaction fingerprints from protein molecular surfaces using geometric deep learning. Nat. Methods 2020, 17, 184–192. [Google Scholar] [CrossRef] [PubMed]
- Isert, C.; Atz, K.; Schneider, G. Structure-based drug design with geometric deep learning. Curr. Opin. Struct. Biol. 2023, 79, 102548. [Google Scholar] [CrossRef]
- Gainza, P.; Wehrle, S.; Van Hall-Beauvais, A.; Marchand, A.; Scheck, A.; Harteveld, Z.; Buckley, S.; Ni, D.; Tan, S.; Sverrisson, F.; et al. De novo design of protein interactions with learned surface fingerprints. Nature 2023, 617, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Porollo, A.; Meller, J. Prediction-Based Fingerprints of Protein–Protein Interactions. Proteins: Struct. Funct. Bioinform. 2007, 66, 630–645. [Google Scholar] [CrossRef] [PubMed]
- Sverrisson, F.; Feydy, J.; Correia, B.E.; Bronstein, M.M. Fast end-to-end learning on protein surfaces. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Virtual Attendance, 20–25 June 2021; pp. 15272–15281. [Google Scholar]
- dMaSIF—Fast end-to-end learning on protein surfaces. Available online: github.com/FreyrS/dMaSIF (accessed on 18 May 2023).
- Reiser, P.; Neubert, M.; Eberhard, A.; Torresi, L.; Zhou, C.; Shao, C.; Metni, H.; van Hoesel, C.; Schopmans, H.; Sommer, T.; et al. Graph neural networks for materials science and chemistry. Commun. Mater. 2022, 3, 93. [Google Scholar] [CrossRef]
- Kriegeskorte, N.; Kievit, R.A. Representational geometry: Integrating cognition, computation, and the brain. Trends Cogn. Sci. 2013, 17, 401–412. [Google Scholar] [CrossRef] [Green Version]
- Cohen, U.; Chung, S.; Lee, D.D.; Sompolinsky, H. Separability and geometry of object manifolds in deep neural networks. Nat. Commun. 2020, 11, 746. [Google Scholar] [CrossRef] [Green Version]
- Kriegeskorte, N. Deep Neural Networks: A New Framework for Modeling Biological Vision and Brain Information Processing. Annu. Rev. Vis. Sci. 2015, 1, 417–446. [Google Scholar] [CrossRef] [Green Version]
- Goyal, A.; Bengio, Y. Inductive biases for deep learning of higher-level cognition. Proc. R. Soc. A 2022, 478, 20210068. [Google Scholar] [CrossRef]
- Wen, H.; Shi, J.; Zhang, Y.; Lu, K.H.; Cao, J.; Liu, Z. Neural Encoding and Decoding with Deep Learning for Dynamic Natural Vision. Cereb. Cortex 2018, 28, 4136–4160. [Google Scholar] [CrossRef]
- Fusi, S.; Miller, E.K.; Rigotti, M. Why neurons mix: High dimensionality for higher cognition. Curr. Opin. Neurobiol. 2016, 37, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Avena-Koenigsberger, A.; Misic, B.; Sporns, O. Communication dynamics in complex brain networks. Nat. Rev. Neurosci. 2018, 19, 17–33. [Google Scholar] [CrossRef] [PubMed]
- Amari, S.I. Information geometry and its applications: Convex function and dually flat manifold. In Proceedings of the Emerging Trends in Visual Computing: LIX Fall Colloquium, Palaiseau, France, 18–20 November 2008; Revised Invited Papers, March 2009. pp. 75–102. [Google Scholar]
- Mannakee, B.K.; Ragsdale, A.P.; Transtrum, M.K.; Gutenkunst, R.N. Sloppiness and the geometry of parameter space. In Uncertainty in Biology: A Computational Modeling Approach; Geris, L., Gomez-Cabrero, D., Eds.; Springer International: Cham, Switzerland, 2016. [Google Scholar]
- Hestness, J.; Narang, S.; Ardalani, N.; Diamos, G.; Jun, H.; Kianinejad, H.; Patwary, M.; Ali, M.; Yang, Y.; Zhou, Y. Deep Learning Scaling is Predictable, Empirically. arXiv 2017, arXiv:1712.00409. [Google Scholar]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Friedman, R. Geometry-Based Deep Learning in the Natural Sciences. Encyclopedia 2023, 3, 781-794. https://doi.org/10.3390/encyclopedia3030056
Friedman R. Geometry-Based Deep Learning in the Natural Sciences. Encyclopedia. 2023; 3(3):781-794. https://doi.org/10.3390/encyclopedia3030056
Chicago/Turabian StyleFriedman, Robert. 2023. "Geometry-Based Deep Learning in the Natural Sciences" Encyclopedia 3, no. 3: 781-794. https://doi.org/10.3390/encyclopedia3030056