Evaluation of Acridine Orange Staining for a Semi-Automated Urinalysis Microscopic Examination at the Point-of-Care
Abstract
:1. Introduction
2. Materials and Methods
2.1. Point-of-Care (POC) Image-Based System
2.2. Human Subject Population
2.3. Clinical Manual Microscopic Examination
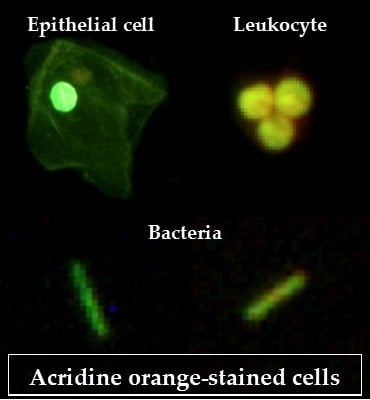
2.4. Acridine Orange (AO) Staining Method
2.5. Automated Image Analysis and Cell Enumeration
2.6. Performance of the Point-of-Care (POC) System Compared to the Clinical Results
3. Results
3.1. Qualitative Imaging Performance
3.2. Colorimetric and Image-Based Morphology Features
3.3. Point-of-Care (POC) Results Compare Favorably to Urinalysis Clinical Standards
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Foxman, B. Epidemiology of urinary tract infections: Incidence, morbidity, and economic costs. Am. J. Med. 2002, 113, 5–13. [Google Scholar] [CrossRef]
- Íñigo, M.; Coello, A.; Fernández-Rivas, G.; Carrasco, M.; Marcó, C.; Fernández, A.; Casamajor, T.; Ausina, V. Evaluation of the SediMax automated microscopy sediment analyzer and the Sysmex UF-1000i flow cytometer as screening tools to rule out negative urinary tract infections. Clin. Chim. Acta 2016, 456, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Carlson, A.L.; Munigala, S.; Russo, A.J.; McMullen, K.M.; Wood, H.; Jackups, R.; Warren, D.K. Inpatient urine cultures are frequently performed without urinalysis or microscopy: Findings from a large academic medical center. Infect. Control Hosp. Epidemiol. 2017, 38, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Subcommittee on Urinary Tract Infection, Steering Committee on Quality Improvement and Management; Roberts, K.B. Urinary tract infection: Clinical practice guideline for the diagnosis and management of the initial UTI in febrile infants and children 2 to 24 months. Pediatrics 2011, 128, 595–610. [Google Scholar] [CrossRef] [PubMed]
- Chaudhari, P.P.; Monuteaux, M.C.; Bachur, R.G. Microscopic Bacteriuria Detected by Automated Urinalysis for the Diagnosis of Urinary Tract Infection. J. Pediatr. 2018, 202, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.L.; Gaido, L. Laboratory diagnosis of urinary tract infections in adult patients. Clin. Infect. Dis. 2004, 38, 1150–1158. [Google Scholar] [CrossRef] [PubMed]
- Simerville, J.A.; Maxted, W.C.; Pahira, J.J. Urinalysis: A comprehensive review. Am. Fam. Physician 2005, 71, 1153–1162. [Google Scholar] [PubMed]
- Becker, G.J.; Garigali, G.; Fogazzi, G.B. Advances in urine microscopy. Am. J. Kidney Dis. 2016, 67, 954–964. [Google Scholar] [CrossRef]
- Winkel, P.; Statland, B.E.; Jorgensen, K. Urine microscopy, an ill-defined method, examined by a multifactorial technique. Clin. Chem. 1974, 20, 436–439. [Google Scholar]
- İnce, F.D.; Ellidağ, H.Y.; Koseoğlu, M.; Şimşek, N.; Yalçın, H.; Zengin, M.O. The comparison of automated urine analyzers with manual microscopic examination for urinalysis automated urine analyzers and manual urinalysis. Pract. Lab. Med. 2016, 5, 14–20. [Google Scholar] [CrossRef] [Green Version]
- Xiang, D.; Cong, Y.; Wang, C.; Yue, J.; Ma, X.; Lu, Y.; Liu, P.; Ma, J. Development of microscopic review criteria by comparison urine flow cytometer, strip and manual microscopic examination. Clin. Lab. 2012, 58, 979–985. [Google Scholar] [PubMed]
- Gangaram, R.; Ojwang, P.; Moodley, J.; Maharaj, D. The accuracy of urine dipsticks as a screening test for proteinuria in hypertensive disorders of pregnancy. Hypertens. Pregnancy 2005, 24, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Lohr, J.A.; Portilla, M.G.; Geuder, T.G.; Dunn, M.L.; Dudley, S.M. Making a presumptive diagnosis of urinary tract infection by using a urinalysis performed in an on-site laboratory. J. Pediatr. 1993, 122, 22–25. [Google Scholar] [CrossRef]
- Van Nostrand, J.D.; Junkins, A.D.; Bartholdi, R.K. Poor predictive ability of urinalysis and microscopic examination to detect urinary tract infection. Am. J. Clin. Pathol. 2000, 113, 709–713. [Google Scholar] [CrossRef] [PubMed]
- Luciano, R.; Piga, S.; Federico, L.; Argentieri, M.; Fina, F.; Cuttini, M.; Misirocchi, E.; Emma, F.; Muraca, M. Development of a score based on urinalysis to improve the management of urinary tract infection in children. Clin. Chim. Acta 2012, 413, 478–482. [Google Scholar] [CrossRef]
- Duong, H.P.; Wissing, K.M.; Tram, N.; Mascart, G.; Lepage, P.; Ismaili, K. Accuracy of Automated Flow Cytometry-Based Leukocyte Counts To Rule Out Urinary Tract Infection in Febrile Children: A Prospective Cross-Sectional Study. J. Clin. Microbiol. 2016, 54, 2975–2981. [Google Scholar] [CrossRef] [PubMed]
- Öztürk, A.; Ginis, Z.; Hanci, T.; Yildiz, Z.; Taşlipinar, M.Y.; Fatma, U.; Öztürk, G.; Yalçindağ, A.; Alay, E.F.; Erden, G. Evaluation of diagnostic performance of routine automated urinalysis and association between urinary tract infection and leukocytosis. Acta Med. 2015, 46, 42–47. [Google Scholar]
- Kalra, O.P.; Raizada, A. Approach to a patient with urosepsis. J. Glob. Infect. Dis. 2009, 1, 57–63. [Google Scholar] [CrossRef]
- Mayo, S.; Acevedo, D.; Quinones-Torrelo, C.; Canós, I.; Sancho, M. Clinical laboratory automated urinalysis: Comparison among automated microscopy, flow cytometry, two test strips analyzers, and manual microscopic examination of the urine sediments. J. Clin. Lab. Anal. 2008, 22, 262–270. [Google Scholar] [CrossRef]
- Kim, S.Y.; Park, Y.; Kim, H.; Kim, J.; Koo, S.H.; Kwon, G.C. Rapid Screening of Urinary Tract Infection and Discrimination of Gram-Positive and Gram-Negative Bacteria by Automated Flow Cytometric Analysis Using Sysmex UF-5000. J. Clin. Microbiol. 2018, 56, e2004–e2017. [Google Scholar] [CrossRef]
- Jiang, T.; Chen, P.; Ouyang, J.; Zhang, S.; Cai, D. Urine particles analysis: Performance evaluation of Sysmex UF-1000i and comparison among urine flow cytometer, dipstick, and visual microscopic examination. Scand. J. Clin. Lab. Invest. 2011, 71, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Chien, T.; Kao, J.; Liu, H.; Lin, P.; Hong, J.; Hsieh, H.; Chien, M. Urine sediment examination: A comparison of automated urinalysis systems and manual microscopy. Clin. Chim. Acta 2007, 384, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Laiwejpithaya, S.; Wongkrajang, P.; Reesukumal, K.; Bucha, C.; Meepanya, S.; Pattanavin, C.; Khejonnit, V.; Chuntarut, A. UriSed 3 and UX-2000 automated urine sediment analyzers vs manual microscopic method: A comparative performance analysis. J. Clin. Lab. Anal. 2018, 32, e22249. [Google Scholar] [CrossRef] [PubMed]
- Zaman, Z. Automated urine screening devices make urine sediment microscopy in diagnostic laboratories economically viable. Clin. Chem. Lab. Med. 2015, 53, s1509–s1511. [Google Scholar] [CrossRef] [PubMed]
- Pai, N.P.; Vadnais, C.; Denkinger, C.; Engel, N.; Pai, M. Point-of-care testing for infectious diseases: Diversity, complexity, and barriers in low-and middle-income countries. PLoS Med. 2012, 9, e1001306. [Google Scholar] [CrossRef] [PubMed]
- Vashist, S.K.; Luppa, P.B.; Yeo, L.Y.; Ozcan, A.; Luong, J.H. Emerging technologies for next-generation point-of-care testing. Trends Biotechnol. 2015, 33, 692–705. [Google Scholar] [CrossRef] [PubMed]
- Schot, M.J.; van Delft, S.; Kooijman-Buiting, A.M.; de Wit, N.J.; Hopstaken, R.M. Analytical performance, agreement and user-friendliness of six point-of-care testing urine analysers for urinary tract infection in general practice. Bmj Open 2015, 5, e006857-2014-006857. [Google Scholar] [CrossRef] [PubMed]
- Ra, M.; Muhammad, M.S.; Lim, C.; Han, S.; Jung, C.; Kim, W. Smartphone-based point-of-care urinalysis under variable illumination. Ieee J. Transl. Eng. Health Med. 2018, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.T.; Li, L.; Zhu, Y.; Bowden, A.K. Low-power, low-cost urinalysis system with integrated dipstick evaluation and microscopic analysis. Lab. A Chip 2018, 18, 2111–2123. [Google Scholar] [CrossRef]
- Melamed, M.R.; Darzynkiewicz, Z.; Traganos, F.; Sharpless, T. Cytology automation by flow cytometry. Cancer Res. 1977, 37, 2806–2812. [Google Scholar]
- Smith, Z.J.; Gao, T.; Chu, K.; Lane, S.M.; Matthews, D.L.; Dwyre, D.M.; Hood, J.; Tatsukawa, K.; Heifetz, L.; Wachsmann-Hogiu, S. Single-step preparation and image-based counting of minute volumes of human blood. Lab. A Chip 2014, 14, 3029–3036. [Google Scholar] [CrossRef]
- Forcucci, A.; Pawlowski, M.E.; Crannell, Z.; Pavlova, I.; Richards-Kortum, R.; Tkaczyk, T.S. All-plastic miniature fluorescence microscope for point-of-care readout of bead-based bioassays. J. Biomed. Opt. 2015, 20, 105010. [Google Scholar] [CrossRef] [PubMed]
- Powless, A.J.; Conley, R.J.; Freeman, K.A.; Muldoon, T.J. Considerations for point-of-care diagnostics: Evaluation of acridine orange staining and postprocessing methods for a three-part leukocyte differential test. J. Biomed. Opt. 2017, 22, 035001. [Google Scholar] [CrossRef] [PubMed]
- Collste, L.G.; Darzynkiewicz, Z.; Traganos, F.; Sharpless, T.K.; Sogani, P.; Grabstald, H.; Whitmore, W.F.; Melamed, M.R. Flow cytometry in bladder cancer detection and evaluation using acridine orange metachromatic nucleic acid staining of irrigation cytology specimens. J. Urol. 1980, 123, 478–485. [Google Scholar] [CrossRef]
- Melamed, M.R.; Traganos, F.; Sharpless, T.; Darzynkiewicz, Z. Urinary cytology automation. Preliminary studies with acridine orange stain and flow-through cytofluorometry. Invest. Urol. 1976, 13, 331–338. [Google Scholar] [PubMed]
- Lauer, B.A.; Reller, L.B.; Mirrett, S. Comparison of acridine orange and Gram stains for detection of microorganisms in cerebrospinal fluid and other clinical specimens. J. Clin. Microbiol. 1981, 14, 201–205. [Google Scholar] [PubMed]
- Mirrett, S.; Lauer, B.A.; Miller, G.A.; Reller, L.B. Comparison of acridine orange, methylene blue, and Gram stains for blood cultures. J. Clin. Microbiol. 1982, 15, 562–566. [Google Scholar] [PubMed]
- Pierzyńska-Mach, A.; Janowski, P.A.; Dobrucki, J.W. Evaluation of acridine orange, LysoTracker Red, and quinacrine as fluorescent probes for long-term tracking of acidic vesicles. Cytom. Part. A 2014, 85, 729–737. [Google Scholar] [CrossRef]
- Hampton, M.B.; Chambers, S.T.; Vissers, M.C.; Winterbourn, C.C. Bacterial killing by neutrophils in hypertonic environments. J. Infect. Dis. 1994, 169, 839–846. [Google Scholar] [CrossRef]
- Schiwon, M.; Weisheit, C.; Franken, L.; Gutweiler, S.; Dixit, A.; Meyer-Schwesinger, C.; Pohl, J.; Maurice, N.J.; Thiebes, S.; Lorenz, K. Crosstalk between sentinel and helper macrophages permits neutrophil migration into infected uroepithelium. Cell 2014, 156, 456–468. [Google Scholar] [CrossRef]
- Veljkovic, K.; Rodriguez-Capote, K.; Bhayana, V.; Pickersgill, R.; Beattie, J.; Clark, L.; Kavsak, P.A. Assessment of a four hour delay for urine samples stored without preservatives at room temperature for urinalysis. Clin. Biochem. 2012, 45, 856–858. [Google Scholar] [CrossRef]
- McHugh, M.L. Interrater reliability: The kappa statistic. Biochem. Med. 2012, 22, 276–282. [Google Scholar] [CrossRef]
- Regeniter, A.; Haenni, V.; Risch, L.; Kochli, H.; Colombo, J.; Frei, R. Urine analysis performed by flow cytometry: Reference range determination and. Clin. Nephrol. 2001, 55, 384–392. [Google Scholar] [PubMed]
- Cohen, R.A.; Brown, R.S. Microscopic hematuria. N. Engl. J. Med. 2003, 348, 2330–2338. [Google Scholar] [CrossRef] [PubMed]



| Clinical Results (Range) | POC System Results (Count) | ||||||
|---|---|---|---|---|---|---|---|
| Subject | Diagnosis | Bacteria | Leukocytes | Epithelial Cells | Bacteria | Leukocytes | Epithelial Cells |
| 1 | Abdominal pain | 0 | 0 | Few (3–10) | 0 | 0 | 0 |
| 2 | Dysuria | Moderate (10–25) | 5–10 | Few (3–10) | 30 | 5 | 1 |
| 3 | Dysuria | Few (3–10) | 0–5 | Few (3–10) | 4 | 0 | 0 |
| 4 | UTI | Moderate (10–25) | 5–10 | Moderate (11–25) | 2 | 13 | 3 |
| 5 | Pelvic Pain | Moderate (10–25) | 5–10 | Few (3–10) | 22 | 6 | 2 |
| 6 | Dysuria | Trace (0–2) | 5–10 | Trace (0–2) | 2 | 6 | 0 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Powless, A.J.; Prieto, S.P.; Gramling, M.R.; Conley, R.J.; Holley, G.G.; Muldoon, T.J. Evaluation of Acridine Orange Staining for a Semi-Automated Urinalysis Microscopic Examination at the Point-of-Care. Diagnostics 2019, 9, 122. https://doi.org/10.3390/diagnostics9030122
Powless AJ, Prieto SP, Gramling MR, Conley RJ, Holley GG, Muldoon TJ. Evaluation of Acridine Orange Staining for a Semi-Automated Urinalysis Microscopic Examination at the Point-of-Care. Diagnostics. 2019; 9(3):122. https://doi.org/10.3390/diagnostics9030122
Chicago/Turabian StylePowless, Amy J., Sandra P. Prieto, Madison R. Gramling, Roxanna J. Conley, Gregory G. Holley, and Timothy J. Muldoon. 2019. "Evaluation of Acridine Orange Staining for a Semi-Automated Urinalysis Microscopic Examination at the Point-of-Care" Diagnostics 9, no. 3: 122. https://doi.org/10.3390/diagnostics9030122