Urinary SARS-CoV-2 RNA Is an Indicator for the Progression and Prognosis of COVID-19
Abstract
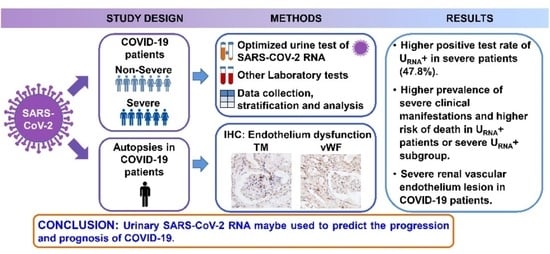
:1. Introduction
2. Materials and Methods
2.1. Study Design and Patients
2.2. Data Collection
2.3. Virological Analysis
2.4. Tissue Sampling and Processing
2.5. Statistical Analysis
3. Results
3.1. Characterization of Patients with URNA+ and URNA−
3.2. Clinical Features and Prognosis of Severe Patients with URNA+ and URNA−
3.3. The Expression of Thrombomodulin (TM) and von Willebrand Factor (vWF) Was Increased in Renal Tissues from Dead COVID-19 Patients
3.4. Theoretical and Mathematic Modeling of Urine Shedding of SARS-CoV-2
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-19. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical characteristics of 138 hospitalized patients with 2019 Novel Coronavirus-infected pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.C.; Cheng, C.H.; Yatsuda, H.; Liu, S.H.; Liu, S.J.; Kogai, T.; Kuo, C.Y.; Wang, R.Y. A novel rapid test to detect Anti-SARS-CoV-2 N protein IgG based on shear horizontal surface acoustic wave (SH-SAW). Diagnostics 2021, 11, 1838. [Google Scholar] [CrossRef] [PubMed]
- Spearman, P. Diagnostic testing for SARS-CoV-2/COVID-19. Curr. Opin. Pediatr. 2021, 33, 122–128. [Google Scholar] [CrossRef]
- Zhang, W.; Du, R.-H.; Li, B.; Zheng, X.-S.; Yang, X.-L.; Hu, B.; Wang, Y.-Y.; Xiao, G.-F.; Yan, B.; Shi, Z.-L.; et al. Molecular and serological investigation of 2019-nCoV infected patients: Implication of multiple shedding routes. Emerg. Microbes Infect. 2020, 9, 386–389. [Google Scholar] [CrossRef] [Green Version]
- Ling, Y.; Xu, S.-B.; Lin, Y.-X.; Tian, D.; Zhu, Z.-Q.; Dai, F.-H.; Wu, F.; Song, Z.-G.; Huang, W.; Chen, J.; et al. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin. Med. J. 2020, 133, 1039–1043. [Google Scholar] [CrossRef]
- Guan, W.-J.; Zhong, N.-S. Clinical characteristics of Covid-19 in China. N. Engl. J. Med. 2020, 382, 1859–1862. [Google Scholar]
- Xie, C.; Jiang, L.; Huang, G.; Pu, H.; Gong, B.; Lin, H.; Ma, S.; Chen, X.; Long, B.; Si, G.; et al. Comparison of different samples for 2019 novel coronavirus detection by nucleic acid amplification tests. Int. J. Infect. Dis. 2020, 93, 264–267. [Google Scholar] [CrossRef]
- Young, B.E.; Ong, S.W.X.; Kalimuddin, S.; Low, J.G.; Tan, S.Y.; Loh, J.; Ng, O.T.; Marimuthu, K.; Ang, L.W.; Mak, T.M.; et al. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA 2020, 323, 1488–1494. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Li, X.; Chen, H.; Yan, S.; Li, D.; Li, Y.; Gong, Z. Coronavirus Disease 19 Infection does not result in acute kidney injury: An analysis of 116 hospitalized patients from Wuhan, China. Am. J. Nephrol. 2020, 51, 343–348. [Google Scholar] [CrossRef]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological assessment of hospitalized patients with COVID-19. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef] [Green Version]
- Newsome, R.C.; Gauthier, J.; Hernandez, M.C.; Abraham, G.E.; Robinson, T.O.; Williams, H.B.; Sloan, M.; Owings, A.; Laird, H.; Christian, T.; et al. The gut microbiome of COVID-19 recovered patients returns to uninfected status in a minority-dominated United States cohort. Gut Microbes 2021, 13, 1–15. [Google Scholar]
- Lo, I.L.; Lio, C.F.; Cheong, H.H.; Lei, C.I.; Cheong, T.H.; Zhong, X.; Tian, Y.; Sin, N.N. Evaluation of SARS-CoV-2 RNA shedding in clinical specimens and clinical characteristics of 10 patients with COVID-19 in Macau. Int. J. Biol. Sci. 2020, 16, 1698–1707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, F.; Yan, L.; Wang, N.; Yang, S.; Wang, L.; Tang, Y.; Gao, G.; Wang, S.; Ma, C.; Xie, R.; et al. Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin. Infect. Dis. 2020, 71, 793–798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronco, C.; Reis, T. Kidney involvement in COVID-19 and rationale for extracorporeal therapies. Nat. Rev. Nephrol. 2020, 16, 308–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Y.; Luo, R.; Wang, K.; Zhang, M.; Wang, Z.; Dong, L.; Li, J.; Yao, Y.; Ge, S.; Xu, G. Kidney disease is associated with in-hospital death of patients with COVID-19. Kidney Int. 2020, 97, 829–838. [Google Scholar] [CrossRef]
- Ng, J.H.; Hirsch, J.S.; Hazzan, A.; Wanchoo, R.; Shah, H.H.; Malieckal, D.A.; Ross, D.W.; Sharma, P.; Sakhiya, V.; Fishbane, S.; et al. Outcomes among patients hospitalized with COVID-19 and acute kidney injury. Am. J. Kidney Dis. 2021, 77, 204–215. [Google Scholar] [CrossRef]
- New Coronavirus Pneumonia Prevention and Control Program, 5th ed.; National Health Commission of China: Beijing, China, 2020.
- Wang, M.; Wu, Q.; Xu, W.; Qiao, B.; Wang, J.; Zheng, H.; Jiang, S.; Mei, J.; Wu, Z.; Deng, Y.; et al. Clinical diagnosis of 8274 samples with 2019-novel coronavirus in Wuhan. MedRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Diao, B.; Wang, C.; Wang, R.; Feng, Z.; Zhang, J.; Yang, H.; Tan, Y.; Wang, H.; Wang, C.; Liu, L.; et al. Human kidney is a target for novel severe acute respiratory syndrome coronavirus 2 infection. Nat. Commun. 2021, 12, 1–9. [Google Scholar] [CrossRef]
- Ye, M.; Wysocki, J.; William, J.; Soler, M.J.; Cokic, I.; Batlle, D. Glomerular localization and expression of angiotensin-converting enzyme 2 and angiotensin-converting enzyme: Implications for Albuminuria in diabetes. J. Am. Soc. Nephrol. 2006, 17, 3067–3075. [Google Scholar] [CrossRef] [Green Version]
- Moura, I.B.; Buckley, A.M.; Wilcox, M.H. Can SARS-CoV-2 be transmitted via faeces? Curr. Opin. Gastroenterol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.I.; Kim, S.G.; Kim, S.M.; Kim, E.H.; Park, S.J.; Yu, K.M.; Chang, J.H.; Kim, E.J.; Lee, S.; Casel, M.A.B.; et al. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe 2020, 27, 704–709. [Google Scholar] [CrossRef] [PubMed]
- Westblade, L.F.; Brar, G.; Pinheiro, L.C.; Paidoussis, D.; Rajan, M.; Martin, P.; Goyal, P.; Sepulveda, J.L.; Zhang, L.; George, G.; et al. SARS-CoV-2 viral load predicts mortality in patients with and without cancer who are hospitalized with COVID-19. Cancer Cell 2020, 38, 661–671. [Google Scholar] [CrossRef] [PubMed]
- Mezoh, G.; Crowther, N. Deciphering endothelial dysfunction in the HIV-infected population. Adv. Exp. Med. Biol. 2019, 1134, 193–215. [Google Scholar] [PubMed]
- Fan, P.-C.; Chang, C.-H.; Chen, Y.-C. Biomarkers for acute cardiorenal syndrome. Nephrology 2018, 23, 68–71. [Google Scholar] [CrossRef] [Green Version]
- Page, A.V.; Liles, W.C. Biomarkers of endothelial activation/dysfunction in infectious diseases. Virulence 2013, 4, 507–516. [Google Scholar] [CrossRef] [Green Version]
- Conway, E.M. Thrombomodulin and its role in inflammation. Semin. Immunopathol. 2012, 34, 107–125. [Google Scholar] [CrossRef] [PubMed]
- Morser, J. Thrombomodulin links coagulation to inflammation and immunity. Curr. Drug Targets 2012, 13, 421–431. [Google Scholar] [CrossRef]
- Kwaifa, I.K.; Bahari, H.; Yong, Y.K.; Noor, S.M. Endothelial dysfunction in obesity-induced inflammation: Molecular mechanisms and clinical implications. Biomolecules 2020, 10, 291. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.-J.; Wu, Z.-Y.; Nie, X.-W.; Bian, J.-S. Role of endothelial dysfunction in cardiovascular diseases: The link between inflammation and hydrogen sulfide. Front. Pharmacol. 2020, 10, 1568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef] [Green Version]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamming, I.; Timens, W.; Bulthuis, M.L.C.; Lely, A.T.; Navis, G.V.; van Goor, H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in under-standing SARS pathogenesis. J. Pathol. 2004, 203, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.K.; Lees, J.S.; Drake, T.M.; Docherty, A.B.; Oates, G.; Hardwick, H.E.; Russell, C.D.; Merson, L.; Dunning, J.; Nguyen-Van-Tam, J.S.; et al. Acute kidney injury in patients hospitalised with COVID-19 from the ISARIC WHO CCP-UK Study: A prospective, multicentre cohort study. Nephrol. Dial. Transplant. 2021. [Google Scholar] [CrossRef] [PubMed]
- Marques, F.; Gameiro, J.; Oliveira, J.; Fonseca, J.A.; Duarte, I.; Bernardo, J.; Branco, C.; Costa, C.; Carreiro, C.; Braz, S.; et al. Acute kidney disease and mortality in acute kidney injury patients with COVID-19. J. Clin. Med. 2021, 10, 4599. [Google Scholar] [CrossRef] [PubMed]
- Moledina, D.G.; Simonov, M.; Yamamoto, Y.; Alausa, J.; Arora, T.; Biswas, A.; Cantley, L.G.; Ghazi, L.; Greenberg, J.H.; Hinchcliff, M.; et al. The association of COVID-19 with acute kidney injury independent of severity of illness: A multicenter cohort study. Am. J. Kidney Dis. 2021, 77, 490–499. [Google Scholar] [CrossRef] [PubMed]
- Chan, L.; Chaudhary, K.; Saha, A.; Chauhan, K.; Vaid, A.; Zhao, S.; Paranjpe, I.; Somani, S.; Richter, F.; Miotto, R.; et al. AKI in hospitalized patients with COVID-19. J. Am. Soc. Nephrol. 2021, 32, 151–160. [Google Scholar] [CrossRef]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef] [Green Version]
- Su, H.; Yang, M.; Wan, C.; Yi, L.-X.; Tang, F.; Zhu, H.-Y.; Yi, F.; Yang, H.-C.; Fogo, A.B.; Nie, X.; et al. Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China. Kidney Int. 2020, 98, 219–227. [Google Scholar] [CrossRef] [PubMed]





| All Patients (n = 53) | Urinary SARS-CoV-2 | p Value | Illness Severity | p Value | |||||
|---|---|---|---|---|---|---|---|---|---|
| URNA− (n = 38, 71.7%) | URNA+ (n = 15, 28.3%) | URNA− vs. URNA+ | Non-Severe | Severe | S URNA+ vs. S URNA− | ||||
| URNA− (n = 26, 86.7%) | URNA+ (n = 4, 13.3%) | URNA− (n = 12, 52.2%) | URNA+ (n = 11, 47.8%) | ||||||
| Demographic characteristic (No., %) | |||||||||
| Age, years | 52 (42.0–66.0) | 51.0 (38.3–59.8) | 61.0 (48.5–72.0) | 0.016 | 50.5 (37.3–56.8) | 50.0 (48.5–60.8) | 55.0 (51.0–63.5) | 66.0 (50.0–72.0) | 0.126 |
| Female | 31/53 (58%) | 22/38 (58%) | 9/15 (60%) | 0.889 | 17/26 (65%) | 3/4 (75%) | 5/12 (42%) | 6/11 (55%) | 0.684 |
| Respiratory symptoms (No., %) | |||||||||
| Fever | 41/53 (77.4%) | 31/38 (73.7%) | 13/15 (86.7%) | 0.969 | 16/26 (61.5%) | 4/4 (100.0%) | 12/12 (100.0%) | 9/11 (81.8%) | 0.217 |
| Cough | 38/53 (71.7%) | 31/38 (73.7%) | 10/15 (66.7%) | 0.421 | 19/26 (73.1%) | 3/4 (75.0%) | 9/12 (75.0%) | 7/11 (63.6%) | 0.667 |
| Sputum production | 13/53 (24.5%) | 9/38 (23.7%) | 6/15 (40.0%) | 0.396 | 5/26 (19.2%) | 3/4 (75.0%) | 2/12 (16.7%) | 3/11 (27.3%) | 0.640 |
| Fatigue | 18/53 (34.0%) | 11/38 (29.0%) | 7/15 (46.7%) | 0.220 | 9/26 (34.6%) | 3/4 (75.0%) | 2/12 (16.7%) | 4/11 (36.4%) | 0.371 |
| Chest tightness | 14/53 (26.4%) | 6/38 (15.8%) | 9/15 (60.0%) | 0.001 | 3/26 (11.5%) | 3/4 (75.0%) | 2/12 (16.7%) | 6/11 (54.6%) | 0.089 |
| Shortness of breath | 19/53 (35.9%) | 12/38 (31.6%) | 10/15 (66.7%) | 0.02 | 0/26 (0.0%) | 0/4 (0.0%) | 9/12 (75.0%) | 10/11 (90.9%) | 0.590 |
| Radiological appearance (No., %) | |||||||||
| Unilateral pneumonia | 2/53 (3.77%) | 2/38 (5.3%) | 0/15 (0.0%) | 0.365 | 2/26 (7.7%) | 0/4 (0.0%) | 0/12 (0.0%) | 0/11 (0.0%) | UTC |
| Bilateral pneumonia | 35/53 (66.0%) | 26/38 (68.4%) | 9/15 (60.0%) | 0.560 | 17/26 (62.4%) | 1/4 (25.0%) | 9/12 (75.0%) | 8/11 (72.7%) | 1.000 |
| Multiple “ground-glass opacity” lesions | 20/35 (37.7%) | 17/38 (44.7%) | 3/15 (20.0%) | 0.094 | 14/26 (53.9%) | 0/4 (0.0%) | 3/12 (25.0%) | 3/11 (27.3%) | 1 |
| Comorbidities (No., %) | |||||||||
| Hypertension | 19/53 (35.9%) | 11/38 (29.0%) | 8/15 (53.3%) | 0.095 | 9/26 (34.6%) | 0/4 (0.0%) | 2/12 (16.7%) | 8/11 (72.7%) | 0.012 |
| Diabetes | 7/53 (13.2%) | 5/38 (13.2%) | 2/15 (13.3%) | 0.986 | 3/26 (11.5%) | 0/4 (0.0%) | 2/12 (16.7%) | 2/11 (18.2%) | 1.000 |
| Cardiovascular diseases | 6/53 (11.3%) | 2/38 (5.3%) | 4/15 (26.7%) | 0.083 | 2/26 (7.7%) | 0/4 (0.0%) | 0/12 (0.0%) | 4/11 (36.4%) | 0.037 |
| Chronic renal disease | 2/53 (3.8%) | 2/38 (5.3%) | 0/15 (0.0%) | 0.365 | 2/26 (7.7%) | 0/4 (0.0%) | 0/12 (0.0%) | 0/11 (0.0%) | UTC |
| In-hospital death (No., %) | 6/53 (11.3%) | 2/38 (5.3%) | 4/15 (26.7%) | 0.083 | 0/26 (0.0%) | 0/4 (0.0%) | 2/12 (16.7%) | 4/11 (36.4%) | 0.371 |
| All Patients (n = 53) | Urinary SARS-CoV-2 | p Value | Illness Severity | p Value | |||||
|---|---|---|---|---|---|---|---|---|---|
| URNA− (n = 38) | URNA+ (n = 15) | URNA− vs. URNA+ | Non-Severe | Severe | S URNA+ vs. S URNA− | ||||
| URNA− (n = 26) | URNA+ (n = 4) | URNA− (n = 12) | URNA+ (n = 11) | ||||||
| Arterial blood gas (No., %) | |||||||||
| PO2 (<100) | 18/53 (34.0%) | 8/38 (21.1%) | 10/15 (66.7%) | 0.002 | 0/26 (0.0%) | 0/4 (0.0%) | 8/12 (66.7%) | 10/11 (90.9%) | 0.317 |
| PO2 (<80) | 13/53 (24.5%) | 6/38 (15.8%) | 7/15 (46.7%) | 0.046 | 0/26 (0.0%) | 0/4 (0.0%) | 6/12 (50.0%) | 7/11 (63.6) | 0.680 |
| PCO2 (>46) | 9/53 (17.0%) | 5/38 (13.2%) | 4/15 (26.7%) | 0.439 | 0/26 (0.0%) | 0/4 (0.0%) | 5/12 (41.7%) | 4/11 (36.4%) | 1.000 |
| SaO2 (≤93) | 23/53 (43.4%) | 12/38 (31.6%) | 11/15 (73.3%) | 0.006 | 0/26 (0.0%) | 0/4 (0.0%) | 12/12 (100.0%) | 11/11 (100.0%) | UTC |
| All Patients (n = 53) | Urinary SARS-CoV-2 | p Value | ||
|---|---|---|---|---|
| URNA− (n = 38) | URNA+ (n = 15) | URNA− vs. URNA+ | ||
| T cell | 724.0 (353.0–1035.0) | 809.9 (495.8–1123.5) | 412.0 (213.5–800.5) | 0.019 |
| <723/mL | 25/53 (47.2%) | 16/38 (42.1%) | 9/15 (60.0%) | 0.240 |
| Th cell | 440.0 (189.0–709.0) | 548.0 (219.0–747.8) | 247.0 (128.5–349.0) | 0.011 |
| <404/mL | 26/53 (49.1%) | 14/38 (36.8%) | 12/15 (80.0%) | 0.005 |
| CRP | 16.6 (5.0–77.3) | 5.0 (5.0–38.7) | 77.3 (23.6–95.9) | 0.022 |
| >10 mg/L (No., %) | 30/53 (56.6%) | 15/38 (39.5%) | 15/15 (100.0%) | 0.001 |
| ALT | 31.0 (18.0–58.0) | 27.0 (16.0–57.5) | 52.0 (29.5–81.5) | 0.029 |
| >50 U/L (No., %) | 22/53 (41.5%) | 12/38 (31.6%) | 10/15 (66.7%) | 0.020 |
| AST | 1.19 (0.88–21.00)) | 1.50 (1.00–24.8) | 0.95 (0.74–1.18) | 0.001 |
| <15 U/L (No., %) | 35/53 (66.0%) | 21/38 (55.3%) | 14/15 (93.3%) | 0.008 |
| >40 U/L (No., %) | 3/53 (5.7%) | 2/38 (5.3%) | 1/15 (6.7%) | 1.000 |
| DBIL | 4.9 (2.7–7.4) | 3.8 (2.2–5.5) | 7.2 (5.0–8.7) | 0.001 |
| >8 mmol/L (No., %) | 16/53 (30.2%) | 7/38 (18.4%) | 9/15 (60.0%) | 0.008 |
| LDH | 276.0 (193.0–458.0) | 206.5 (166.3–314.5) | 443.0 (329.5–587.0) | 0.001 |
| >250 U/L (No., %) | 29/53 (54.7%) | 14/38 (36.8%) | 15/15 (100.0%) | 0.001 |
| BUN | 5.2 (4.0–8.8) | 5.0 (3.7–6.4) | 8.8 (4.1–11.7) | 0.032 |
| >8 mmol/L (No., %) | 19/53 (35.8%) | 9/38 (23.7%) | 10/15 (66.7%) | 0.003 |
| eGFR | 102.1 (86.3–115.6) | 103.7 (93.6–119.5) | 82.1 (63.7–99.2) | 0.002 |
| <90 mL/min/1.73 m2 (No., %) | 16/53 (30.2%) | 5/38 (13.2%) | 11/15 (73.3%) | 0.001 |
| Authors | Sampling Method | Detecting Method | Positive Rate | Target Gene | Detection Kit | Participants Condition | Refs. |
|---|---|---|---|---|---|---|---|
| Huiming Wang, et al. | Urine sediments | RT-PCR | 28.3% | NP and ORF1ab | Zhongzhi, Wuhan | 30 non-severe 23 severe | This article |
| Luwen Wang, et al. | Urine sediments | RT-PCR | 7.5% | NP and ORF1ab | Zhongzhi, Wuhan | 48 non-CKD, 5 CKD | [11] |
| Chaolin Huang, et al. | Urine | RT-PCR | 11% | NP and ORF1ab | ND | 9 moderates | [2] |
| Hongzhou Lu, et al. | Urine | RT-PCR | 6.9% | NP and ORF1ab | Master Biotechnology, China | Recovered | [7] |
| Zhenglin Yang, et al. | Urine | RT-PCR | 0% | NP and ORF1ab | GeneoDx (GZ-TRM2, China), Maccura (Sichuan, China) and Liferiver (W-RR-0479-02, China) | 5 Uncomplicated, 14 complicated | [9] |
| Barnaby Edward Young, et al | Urine | RT-PCR | 0% | N, S, and ORF1ab | EZ1 virus mini kit v2.0 (Qiagen) | 6 mild, 4 severe | [10] |
| Roman Wölfel, et al. | Urine | RT-PCR | 0% | E- and RdRp | Tib-Molbiol, Berlin, Germany | mild | [12] |
| Chin Ion Lei, et al. | Urine | qRT-PCR | 0% | NP and ORF1ab | BioGerm, China | 2 mild, 4 moderates | [12] |
| Fujie Zhang, et al. | Urine | RT-PCR and ddPCR | 0% | NP and ORF1ab | Shanghai BioGerm Medical Technology Co. LTD, China (RT-PCR) TargetingOne, Beijing, China (ddPCR) | ND | [13] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Tian, M.; Song, Y.; Liang, W.; Li, X.; Tong, Y.; Wang, H. Urinary SARS-CoV-2 RNA Is an Indicator for the Progression and Prognosis of COVID-19. Diagnostics 2021, 11, 2089. https://doi.org/10.3390/diagnostics11112089
Zhang L, Tian M, Song Y, Liang W, Li X, Tong Y, Wang H. Urinary SARS-CoV-2 RNA Is an Indicator for the Progression and Prognosis of COVID-19. Diagnostics. 2021; 11(11):2089. https://doi.org/10.3390/diagnostics11112089
Chicago/Turabian StyleZhang, Lu, Maoqing Tian, Yuan Song, Wei Liang, Xiaogang Li, Yongqing Tong, and Huiming Wang. 2021. "Urinary SARS-CoV-2 RNA Is an Indicator for the Progression and Prognosis of COVID-19" Diagnostics 11, no. 11: 2089. https://doi.org/10.3390/diagnostics11112089